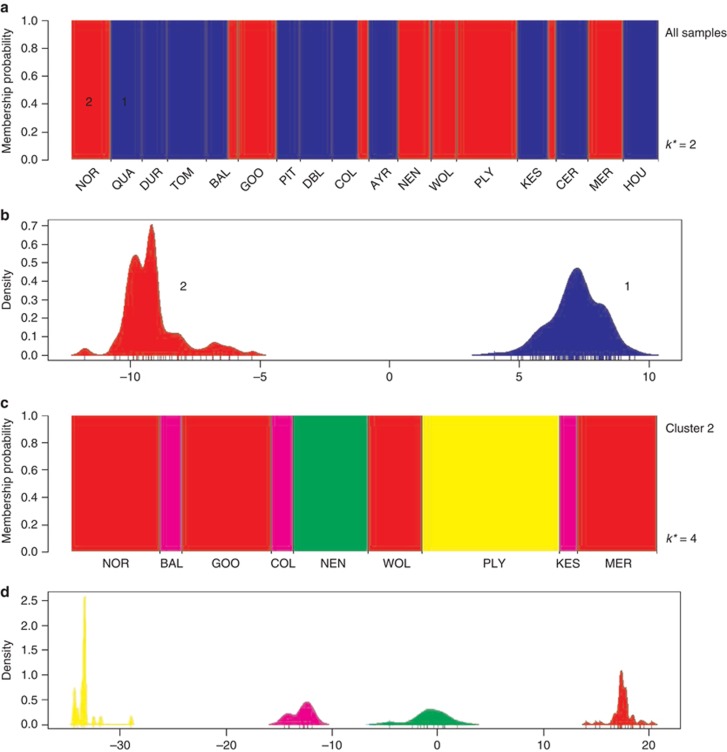
Figure 2.
Results of the DAPCs showing the genetic clustering of 300 Mimulus spp. individuals from 17 populations across Great Britain. (a) Individual probability of membership to either of two genetic clusters (k*=2) identified in the DAPC analysis. Cluster 1 (blue) is associated with diploid individuals (M. guttatus), whereas cluster 2 (red) represents polyploid individuals (M. x robertsii and M. luteus s.l.). The x axis shows the population of origin from north to south, with population codes as given in Table 4. (b) Discriminant analysis using the first 15 principal components shows a clear ability to distinguish the two genetic clusters. The x axis shows the values of the first discriminant function for each individual (vertical bars), and the y axis indicates the smoothed density of observations. (c, d) Analysis of cluster 2 samples only. (c) Individual probabilities of membership to either of four genetic clusters (k*=4) identified in the DAPC analysis. Notice that the tetraploid individuals (M. luteus s.l.) from population COL are not uniquely separated from triploid (M. x robertsii) individuals in other populations (BAL and KES). (d) Discriminant analysis of cluster 2 samples showing the values of the first discriminant function. Colours correspond to the four genetic clusters in (c).