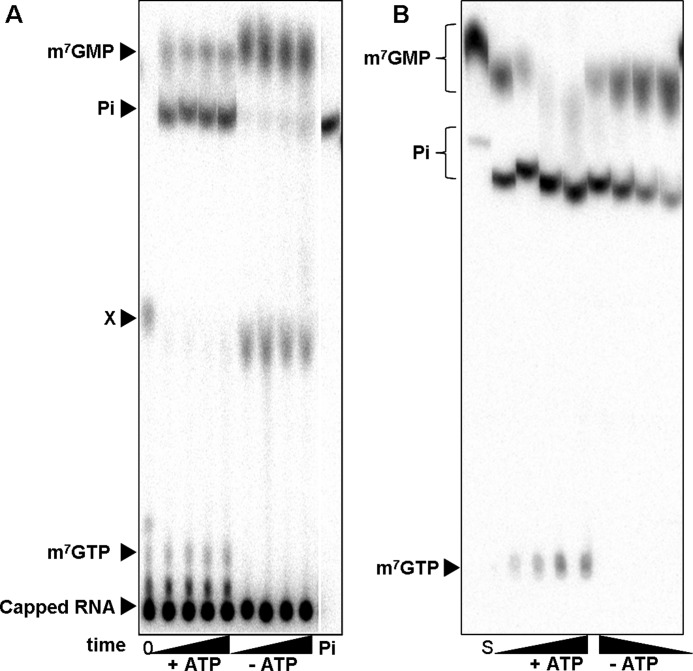
FIGURE 1.
A, shown is release of phosphate during incubation of 32P-labeled capped RNA with Drosophila embryo extract. The reaction mixtures containing 20% embryo extract were preincubated for 10 min with hexokinase and glucose for ATP depletion or complemented with creatine kinase and creatine for ATP regeneration as indicated. The reactions were started by the addition of the 32P-labeled capped RNA (200 nm). At different time points (0, 5, 10, 20, 40 min) 2-μl aliquots were removed and mixed with EDTA. The reaction products were separated by TLC and detected by autoradiography. For details, see “Experimental Procedures.” The first lane contains the substrate at time point 0. Migration of standards is indicated on the left. Labeled m7GMP and m7GpppG were generated by digestion of 32P-labeled capped RNA with P1 nuclease with or without DcpS, respectively. M7GTP was an unlabeled standard detected by UV absorbance, and 32P-orthosphosphate, shown in the last lane, was obtained commercially. All standards were analyzed after the addition to a reaction mixture lacking labeled RNA. An unidentified compound is labeled X. B, release of phosphate during incubation of 32P-labeled m7GMP with Drosophila embryo extract is shown. The reaction mixtures contained 20% embryo extract. ATP-depleting or -regenerating conditions were as in A. The reactions were started by the addition of 32P-labeled m7GMP (200 nm). After 0, 5, 10, 20, and 40 min, 2 μl were removed, and the reactions were stopped with EDTA. Products were separated by TLC and detected by autoradiography. The first lane represents the substrate m7GMP analyzed directly from water. Its migration is thus different from that of samples taken from the reaction mixture. Standards indicated on the left were analyzed from a reaction mixture.