FIGURE 1.
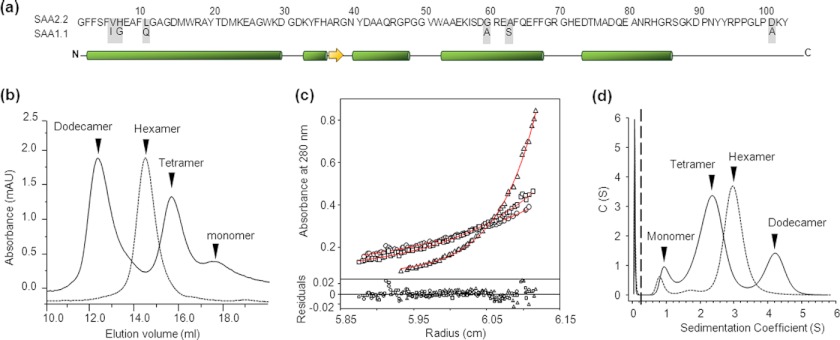
The in vitro oligomeric structure of refolded SAA1.1 and SAA2.2. a, SAA1.1 and SAA2.2 share a high degree of structural similarity, including 94% sequence identity, and similar α-helix and β-sheet secondary structure according to a secondary structure prediction algorithm. b, analytical size-exclusion chromatography shows that SAA2.2 forms a hexamer (elution volume ∼14.7 ml), whereas SAA1.1 folds into a heterogeneous mixture of dodecamer (∼12.4 ml), tetramer (∼15.8 ml), and monomer (∼17.8 ml). mAU, milliabsorbance units. c, shown is equilibrium sedimentation AUC analysis of SAA1.1. Solid lines represent the global fit of representative traces at 10,000 (○), 12,000 (□), and 19,000 rpm (▵). The lower panel shows the residuals for each fit. The Mr obtained from the fit was 61.7 ± 0.9. d, sedimentation velocity AUC experiments showed at least three different sedimentation boundaries for SAA1.1 consistent with a mixture of dodecamer, tetramer, and monomer. SAA2.2 was analyzed in parallel experiments, and the data revealed one major boundary consistent with the hexameric structure observed by SEC and previous AUC experiments (8). All experiments were carried out at 4 °C.