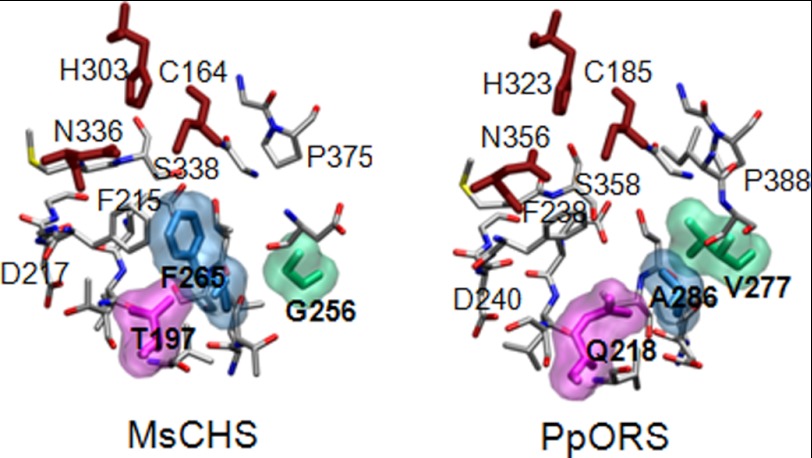
FIGURE 4.
Comparison of the active sites of MsCHS (Protein Data Bank ID code 1bi5) and the PpORS model. The Cys-His-Asn catalytic triads are shown in brick red. The PpORS specific active site residues, Gln218, Val277, and Ala286, are shown as space-filling model with the corresponding Thr197, Gly211, and Gln212 residues in MsCHS. According to the MolProbity parameters (43), the modeled PpORS structure was found to be in the 69th percentile of overall quality (MolProbity score 2.12) compared with all known crystal structures. The model achieved the following scorings: 1.75% for Ramachandran outliers, 8.06% for poor rotamers, and 1.47 for Clashscore.