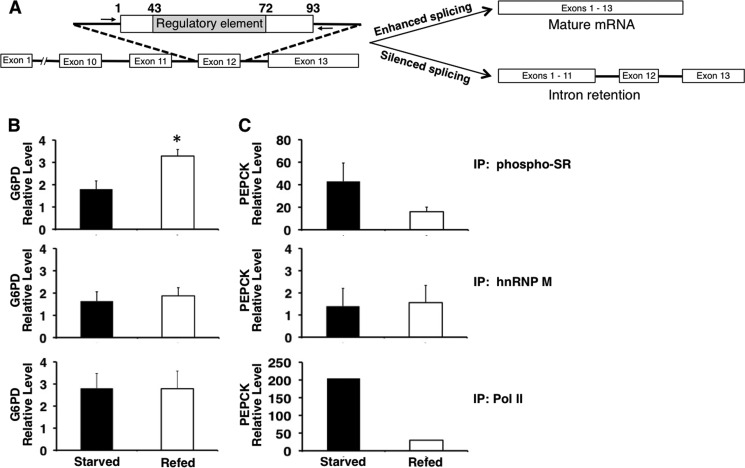
FIGURE 2.
Refeeding increases the binding of phosphorylated SR proteins to the regulatory element of G6PD exon 12 in vivo. ChIP was performed in livers of mice that were starved for 16 h or starved and then refed a high carbohydrate diet for 12 h. A, scheme of the G6PD RNA containing the regulatory element and observed changes in splicing. The arrows denote the location of the primers. The forward primer ends 6 nt upstream of the 3′ splice site of intron 11, and the reverse primer starts 3 nt downstream of the 5′ splice site in intron 12. B, immunoprecipitation (IP) used antibodies that detect: phosphorylated SR protein (phospho-SR), hnRNP M, and RNA polymerase II. The phospho-SR antibody (1H4) detects SRSF1, SRSF2, SRSF3, SRSF4, SRSF5, and SRSF6. The immunoprecipitated DNA was measured using real time PCR and primers adjacent to exon 12 (A). The amount of DNA detected is expressed relative to the input chromatin as described under “Experimental Procedures.” The asterisk indicates p < 0.05, n = 3 mice. C, detection of PEPCK in the immunoprecipitated samples. PEPCK DNA was measured using real time PCR and primers to exon 2 (n = 2–3 mice).