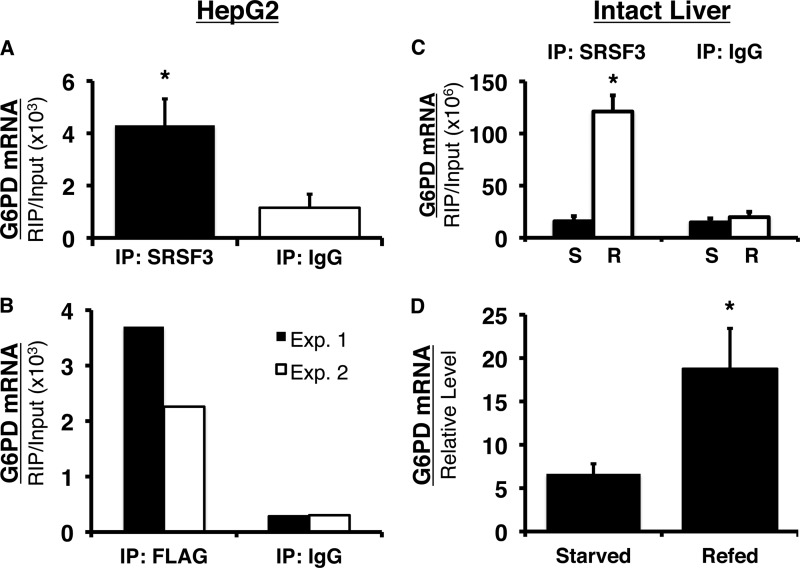
FIGURE 7.
SRSF3 specifically binds to the splicing regulatory element in vivo and refeeding enhances the binding of SRSF3 to the splicing regulatory element in mouse liver. A, HepG2 cells were cross-linked with formaldehyde, and SRSF3 was immunoprecipitated from the lysates. G6PD RNA bound to SRSF3 was detected using primers, which amplify the region between nucleotides 11 and 85 in exon 12 (the regulatory element spans nucleotides 43–72). Immunoprecipitation (IP) with IgG was used as control for nonspecific binding to antibody. The amount of RNA detected is expressed relative to the input RNA in each sample. The data are the means ± S.E. of four separate immunoprecipitations. B, HepG2 cells were transiently transfected with FLAG-tagged SRSF3. The cells were chemically cross-linked 48 h after transfection, and SRSF3 was immunoprecipitated with anti-FLAG antibody. The results of two separate experiments are shown. C, mice were either starved for 18 h or starved for 18 h and refed a high carbohydrate diet for 12 h. RIP was performed on the chemically cross-linked livers using an antibody against SRSF3 or IgG. G6PD RNA immunoprecipitated with SRSF3 or IgG was detected with real time RT-PCR and primers to the region of exon 12 (nucleotides 18–89) that contains the splicing regulatory element (nucleotides 43–72). The amount of detected RNA is expressed relative to the input RNA in each sample. The data are the means ± S.E. of three mice/group. D, total RNA was isolated from a portion of each liver used in the RIP analysis. G6PD mRNA was measured using real time RT-PCR as previously described. Each bar is the mean ± S.E. of three mice. The asterisk indicates a significant difference (p < 0.05).