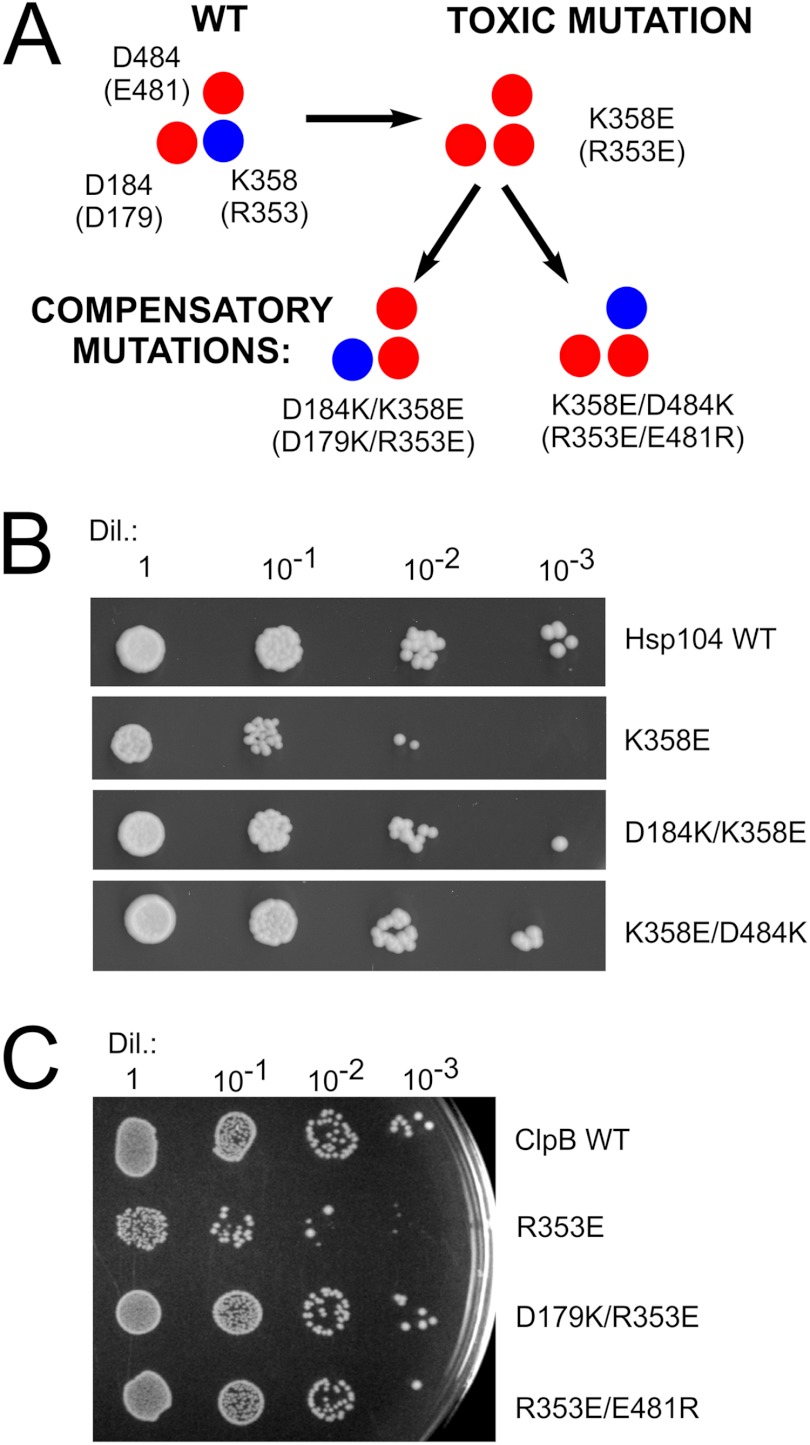
FIGURE 10.
Compensatory mutations, which potentially recreate ionic interactions, suppress the toxic effect observed for both HSP104 and clpB mutants. A, schematic representation of the analyzed amino acids in Hsp104 and (ClpB) variants is shown. B, drop-tests of yeast W303hsp104Δ strain transformed with the pRS313 plasmid carrying variants of the HSP104 gene are shown. Equal amounts of yeast cells (106 cells/ml) were serially diluted (Dil.) and spotted onto the SC (−His) plate. C, shown are drop-tests of E. coli MC4100 ΔclpB strain transformed with the pGB2 plasmid carrying variants of the clpB gene. Equal amounts of cells were serially diluted and spotted onto the plate and grown at 4 °C.