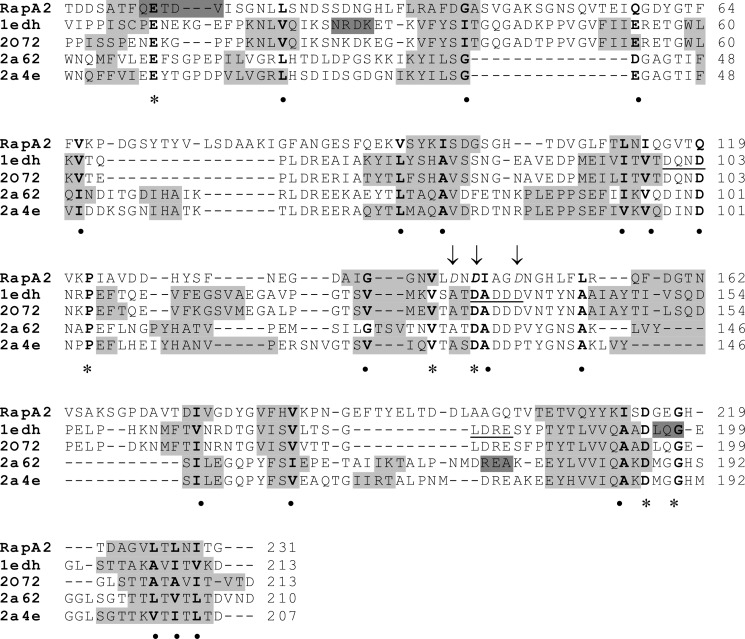
FIGURE 1.
Structural alignment of RapA2 and eukaryotic cadherins. The amino acid sequence of RapA2 was aligned to template sequences according to the MetaServer predictions. The template structures and their PDB entries were: ectodomains 1 and 2 (EC1–2) of mouse E-cadherin (PDB code 1edh), human E-cadherin EC1–2 (PDB 2o72), mouse cadherin-8 EC1–3 (PDB code 2a62), and mouse cadherin-11 EC1–2 (PDB code 2a4e). The predicted RapA2 secondary structure is from SWISS-MODEL, and the template structures were from the Protein Data Bank. Secondary structure elements are shown as a gray background (light gray for β-strands, dark gray for α-helices). Conserved residues are in bold, asterisks denote identical residues, and circles indicate similar residues. The conserved calcium binding motifs in cadherins (DXNDN, DXD and LDRE) are underlined in the sequence of PDB code 1edh. The acidic amino acids of RapA2 mutated to alanine are shown in italics and indicated with arrows.