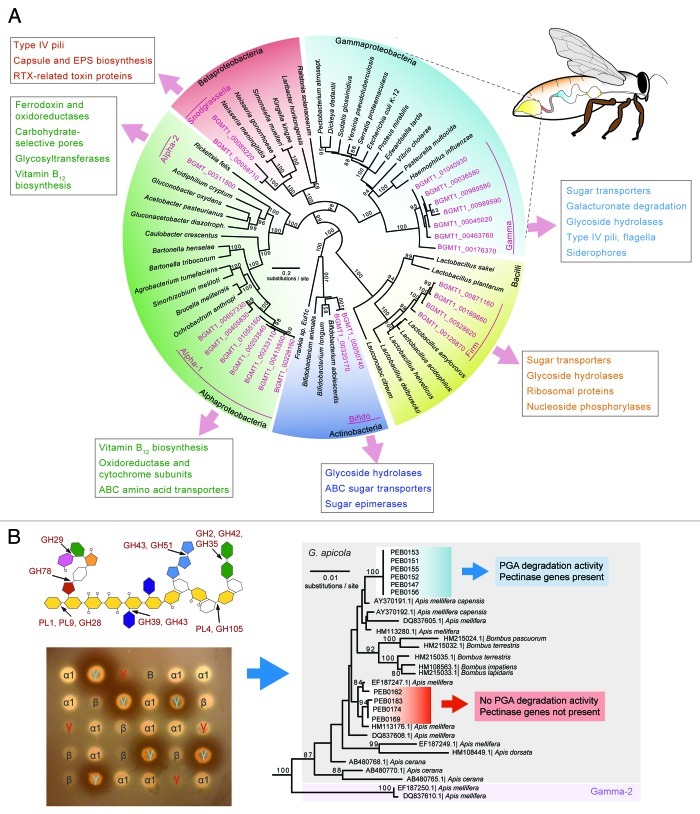
Figure 1. (A) Sequences from the honey bee gut metagenome (pink color) fall into a few phylogenetic lineages distinct from other bacteria. Although there can be more than one species per lineage (Gamma and Firm), the presence of several distinct sequences per lineage indicates genetic diversity within these bee-specific bacteria. Functions belonging to COG categories enriched in one lineage relative to the others are shown in boxes. Colors in the digestive tract of the bee illustration match colors of the different bacterial phyla in the tree and indicate regions where the corresponding bacteria are most abundant.27,48 (Localization of the two Alphaproteobacteria and the Bifidobacterium are not known.) (B) Genetic differences are linked to functional differences, as shown for the example of pectin degradation by G. apicola. The scheme of the pectin molecule shows glycoside hydrolase (GHs) and polysaccharide lyase (PLs) families identified in the honey bee metagenome with their respective cleavage sites. Different colors represent different sugar molecules. Degradation of polygalacturonate (PGA), the major component of pectin, by a subset of G. apicola isolates is demonstrated by clearance zones forming around bacterial patches on an agar plate. γ, Gilliamella; β, Snodgrassella; α1, Alpha-1; and B, Bifido. G. apicola isolates differing in capacity to degrade pectin (depicted by blue and red coloring) belong to distinct phylogenetic clades as shown by a 16S rRNA gene tree. Pectinase genes were absent in isolates negative for PGA degradation.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.