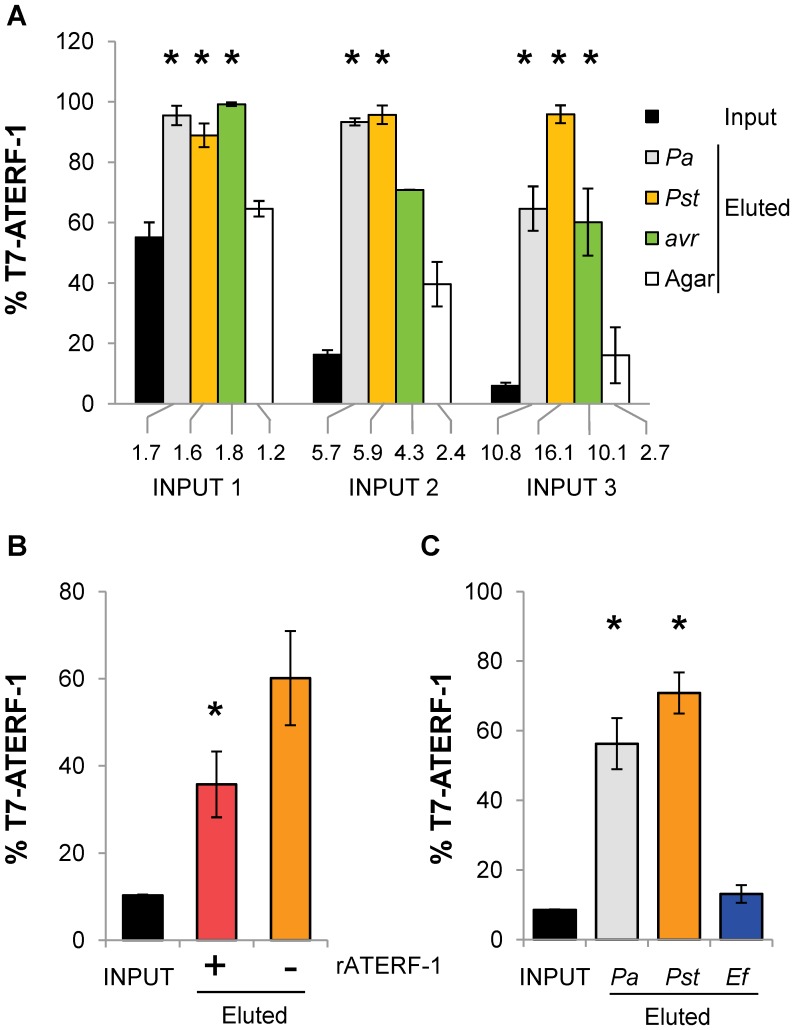
Figure 3. Competitive bio-panning assay shows specific binding of T7-ATERF-1 clone to the three strains of Pseudomonas.
(A) Competition between T7-ATERF-1 phage (which expresses ATERF-1 polypeptide) and T7-C1 (competing phage randomly selected). Mixtures of phage containing 1∶1 (input 1), 1∶6 (input 2) and 1∶17 (input 3) proportions of T7-ATERF-1:T7-C1 clones were prepared to a final concentration of 6.3×109 pfu/ml. Phage mixtures were panned against 1 OD of Pa, Pst, Pst(avrRpt2) (avr) or agarose (Agar) under the same conditions used for T7LAtPa and T7LAtPs libraries. The bar series show the percentage of T7-ATERF-1 clone in the input and in the eluates that were recovered after one round of selection. The numbers below the bars represent the fold enrichment for T7-ATERF-1, calculated as the ratio between eluate and input percentages. Asterisks indicate significant differences (t-test, p<0.05) respect to the agarose (non-specific binding) control. (B) Addition of rATERF-1 protein inhibits binding of T7-ATERF-1 phage to Pst cells. Bacteria were incubated with 10 µg of rATERF-1 (+) or rLacZ (−) during panning with a 1∶10 mixture (input) of T7-ATERF-1:T7-C1 phage. Recovery of T7-ATERF-1 in the eluates was significantly decreased (p<0.05) when rATERF-1 was added to the cells. (C) Competition between T7-ATERF-1 and T7-C1 clone for binding to Pa, Pst or Enterococcus faecalis (Ef). The percentage of T7-ATERF-1 phage recovered from bio-panning with the Gram(+) bacterium was not significantly different from the input (p = 0.1835), whereas eluates recovered from Pseudomonas sp. bio-pannings were enriched in T7-ATERF-1 clone (Pa p = 0.0086 and Pst p = 0.0021).