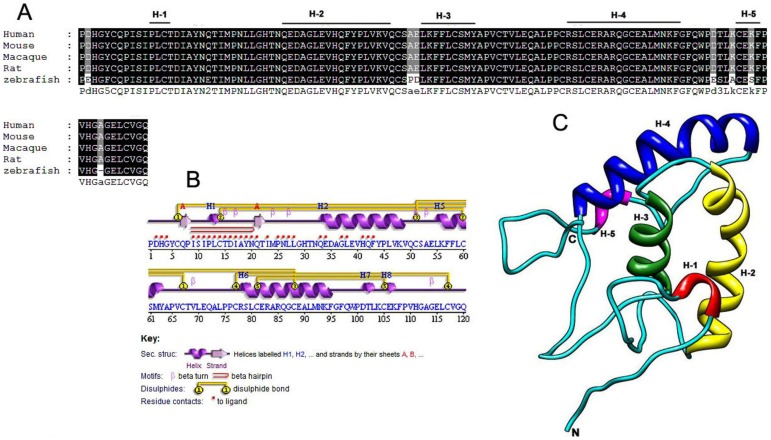
Figure 4. Multiple sequence alignment of orthologous cysteine rich domain. (A).
Conservation of a Frizzled-1 CRD sequence in Human compared with the sequences of Macaque, Mouse, Rat and Zebra-fish. Except a few variations in zebra-fish sequence, the rest of the sequences of CRD are 100% identical. The multiple sequence alignment is generated by ClustalX and labeled by GenDoc. It also contains the important secondary structures elements. (B) Secondary structure of FZD-1 CRD. The secondary elements are predicted by PSIPRED. The strong extensive disulphide bridge formation is the most important feature of CRD. Additionally, beta hairpin motif and important residues for ligand binding are also predicted by PSIPRED. (C) Homology modeled three dimensional structure of CRD. Structure is built by Modeller9v8 using mouse CRD as a template (1IJY). The RMSD value calculated after superimposition was 0.62 Å.