Figure 1.
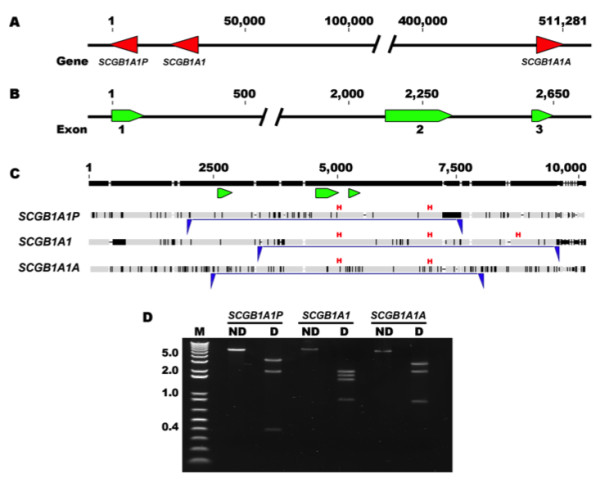
Schematic representation of SCGB1A1 sequencing strategy and enzymatic restriction analysis. (A) The partial genomic sequence of Equus caballus chromosome 12 (based on EquCab2.0, NW_001867370.1) encompassing the three predicted equine SCGB1A1 genes (red triangles). SCGB1A1P and SCGB1A1 are in reverse orientation, while SCGB1A1A is in forward orientation. The chromosome 12 region (original bases 2,788,223 to 3,299,503) includes 511,281 bp bordered by the SCGB1A1P stop codon guanine (position 1) and the SCGB1A1A stop codon guanine (position 511,281). SCGB1A1P is most proximal to the centromere. (B) Predicted structure of an individual SCGB1A1 gene, each containing approximately 2,650 bp, including 3 small exons (green arrows) and 2 introns. (C) Multiple sequence alignment of the three predicted SCGB1A1 genes. The LR-PCR amplification strategy is outlined in blue. Each primer (blue triangle) is specific for a single copy enabling amplification of three distinct PCR products (blue line). Exons are displayed in green under the consensus sequence (black). (D) Restriction enzyme digestion analysis. Non-digested amplicons for SCGB1A1P, SCGB1A1 and SCGB1A1A were 5,559, 6,029 and 5,442 bp in size, respectively. Upon HindIII digestion, SCGB1A1P was detected by 3166, 1942 and 464 bp, SCGB1A1 by 1942, 1,717, 1563, and 891 bp, and SCGB1A1A by 2675, 1942, and 899 bp fragments. H (red), HindIII restriction site; M, 1 Kb + DNA ladder; ND, Non-digested; D, Digested.
