Figure 5.
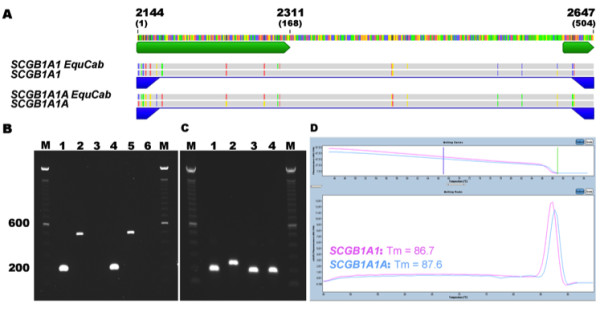
SCGB1A1 and SCGB1A1A qRT-PCR strategy. (A) Multiple sequence alignment of the partial predicted SCGB1A1 and SCGB1A1A EquCab2.0 genomic sequences as well as the SCGB1A1 and SCGB1A1A sequences derived in this study. Two gene-specific primer sets (blue arrows) were designed at sites with the greatest amount of sequence variation (colored annotations above primers). Forward primers are positioned in exon 2 (long green arrow) and reverse primers in exon 3 (small green arrow). This region covers position 2,144 to 2,647 of SCGB1A1 genes corresponding to a 504 bp genomic DNA section (brackets). (B) As expected, SCGB1A1 was detected as a 200 bp PCR product using lung cDNA as a template (Lane 1), a 504 bp band using genomic DNA as template (Lane 2), and undetected with template omission (negative control, Lane 3). Similar findings were noted for SCGB1A1A (Lane 4, 5, and 6, respectively). (C) qRT-PCR products were analyzed after electrophoresis to assure cDNA quality and confirm the absence of genomic DNA contamination. Bands were expected at 203 bp for equine 18S (Lane 1), 254 bp for GAPDH (Lane 2), 200 bp for SCGB1A1 (Lane 3) and SCGB1A1A (Lane 4). (D) Melting curve analysis shows distinct SCGB1A1 and SCGB1A1A melting temperatures of 86.7°, and 87.6° respectively, indicating gene-specific amplification. M, 100-bp marker.
