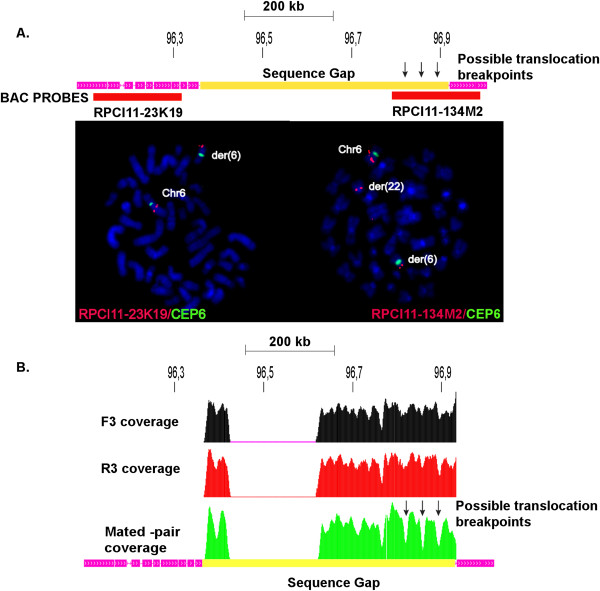
Figure 3.
Translocation breakpoint analysis. A. FISH hybridization detecting the translocation breakpoint. BAC RP11-23 K19 hybridizes to chromosome 6 centromeric of the translocation breakpoint, while RP11-134 M2 hybridizes to both the derivative chromosomes 6 and 22, and encompasses the translocation breakpoint situated within the sequence gap. B. Sequence gap analysis using chimpanzee sequence. Reads from the TS patient were mapped to the chimpanzee genome in order to find candidate regions for the translocation within the human reference gap. In this comparison, we utilized reads where one or both ends mapped to the simian genome. If both ends mapped, we represent the number of mate-pairs spanning across each bin in the region as the green histogram. If only the forward end (i.e. the end closest to the bead) mapped, we show the frequency of mappings as the black histogram. If only the reverse end (most distant from the bead) mapped, we show the frequency as the red histogram. Thus, we can compare the number of mappings in the green histogram, which represent similar arrangements in chimp and human, to the black and green histograms which represent boundaries between shared and non-shared sequences between human and chimp. Areas with low mate-pair coverage are potential sites for the translocation breakpoint and are indicated with arrows.