Abstract
Next-generation sequencing technologies can now be used to directly measure heritable de novo DNA sequence mutations in humans. However, these techniques have not been used to examine environmental factors that induce such mutations and their associated diseases. To address this issue, a working group on environmentally induced germline mutation analysis (ENIGMA) met in October 2011 to propose the necessary foundational studies, which include sequencing of parent–offspring trios from highly exposed human populations, and controlled dose–response experiments in animals. These studies will establish background levels of variability in germline mutation rates and identify environmental agents that influence these rates and heritable disease. Guidance for the types of exposures to examine come from rodent studies that have identified agents such as cancer chemotherapeutic drugs, ionizing radiation, cigarette smoke, and air pollution as germ-cell mutagens. Research is urgently needed to establish the health consequences of parental exposures on subsequent generations.
Keywords: Germ cell, Heritable mutation, Next generation sequencing, Copy number variants
1. Commentary
Over 12,000 genes are identified in the Online Mendelian Inheritance in Man database (OMIM: www.ncbi.nlm.nih.gov/ omim/); sequence variants in these genes are associated with a diverse array of genetic phenotypes. De novo mutations occur in each generation and are increasingly being recognized as contributing to a range of human disorders associated with a broad spectrum of these genes, including autism, schizophrenia, intellectual disability, and epilepsy [1–5]. Recent estimates suggest that a human genome on average contains approximately 100 loss-of-function variants, with as many as 20 exhibiting complete loss of gene function [6]. However, despite major advances in human genomics, the causative variables that influence mutational load have remained an enigma. Although animal models have demonstrated that exposure to mutagens in drugs, food, and the environment can cause germ-cell mutations, no agent has been shown definitively to produce heritable mutations in humans despite the similarity in gametogenic processes in humans and rodents. For example, of the47 agents tested to date for germ cell mutagenesis, 33 chemicals, ionizing radiation, and 3 complex mixtures (urban air, and main- and side-stream cigarette smoke) cause germ-cell mutations in rodents [7–13]. It is likely that some of these agents induce germ-cell mutations in sufficiently exposed humans [14].
Identifying environmentally induced human germline mutations has been stymied by a lack of technologies that can efficiently and economically provide a high-resolution map of a large number of individual genomes [15–17]. Thus, the ENIGMA working group met recently1 to assess the current state of knowledge in environmental mutagenesis and the potential role of environmental agents in causing germline mutations and sporadic disease. The goal was to address how genomic technologies might be deployed to identify variables that impact germline mutation rates and heritable epigenetic changes in humans. The group developed a vision for future research that focuses on studies designed to identify environmental factors that may influence de novo mutation rates and sporadic disease traits in individuals and populations.
2. Opportunities and challenges
Two recent studies of human families directly characterized and quantified de novo DNA mutations derived from both male and female gametes [18,19]. Sequence and structural differences were identified by comparing whole genome sequences or copy number variants between offspring and their parents. These findings show that direct estimates of de novo mutation rates are comparable to indirect estimates from population studies [20,21] and suggest an average mutation rate of 1 × 10−8 per nucleotide per gamete per generation for single nucleotide variants, and 3 × 10−2 per genome per generation for copy number variants greater than 500 bp [5]. Variation in the numbers of copies of a particular region of DNA has generated great excitement not only because of its appreciable rate of occurrence, but also because of its likely impact on human health [22]. In addition to sequencing parents and their offspring, emerging technologies may soon permit quantification of DNA mutation frequencies directly in sperm [23], which will open up new avenues of research in this field. The power of such pedigree and sperm analyses seems clear, presenting tremendous new opportunities for research on the factors influencing mutation rates, such as age, sex, and social and environmental factors. In particular, the study of environmental factors contributing to variation in mutation frequency is a key opportunity. Human cohorts exposed to xenobiotic agents were identified as priority study subjects because extensive animal data, and some human data, support the contention that these exposures should induce heritable DNA changes. Examples include individuals receiving radio- or chemotherapy before conceiving children [12,24], inhabitants of radioactively contaminated areas (e.g., near Chernobyl) [25], cigarette smokers [8,26,27], sickle cell anemia patients treated with hydroxyurea [28], and populations exposed to elevated levels of air pollution [11,29].
A critical first step to advance human germ-cell biology will be to precisely establish the baseline of human germline genetic and epigenetic variability. The range of variation in mutation frequency among gametes and individuals is quite large [18] and represents the background against which any induced mutations must be assessed. Consequently, it is difficult to estimate the needed power of studies over a range of anticipated effect sizes.
An additional challenge is minimizing the error rates associated with next-generation sequencing that can lead to an excess of false positives [17]. However, adequate genomic coverage and sample size, in combination with statistical modeling and the increasing ease with which mutations can be validated greatly diminish the number of false positives [19]. In addition, new technologies are emerging that may overcome this limitation by substantially reducing sequencing error rates [30]. Thus, technological advances make quantifying rates and spectra of inherited mutations in the offspring of exposed individuals an achievable goal.
3. Vision and recommendations
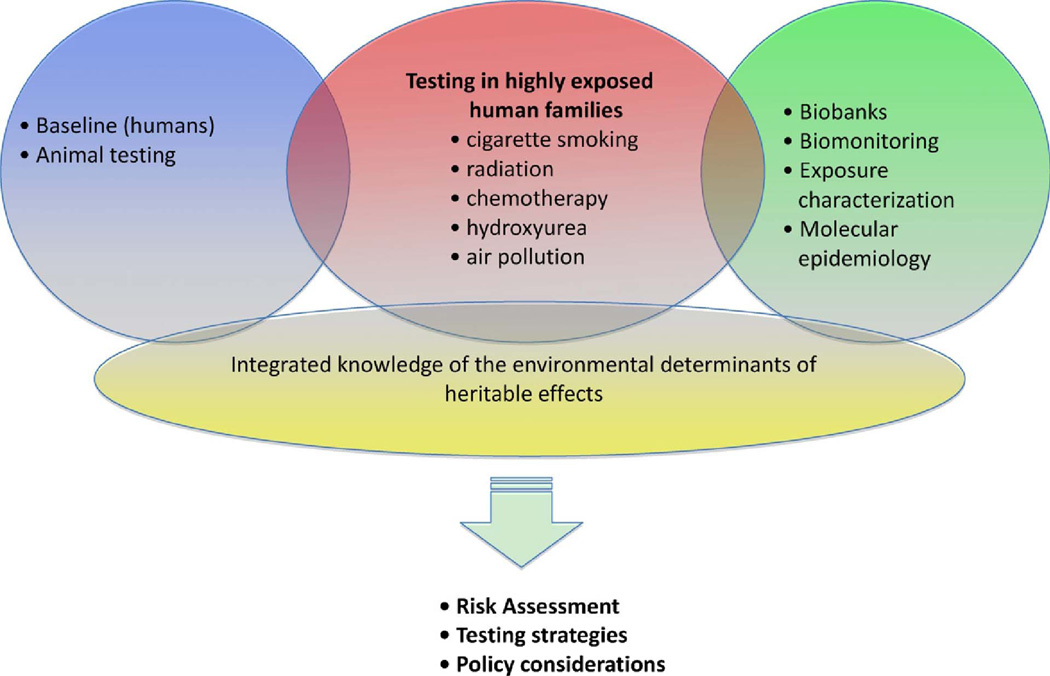
The technologies necessary to discover the complex variables that influence germline genome stability are largely available [17]. Given this premise, the ENIGMA working group recommends a tiered plan comprised of a combination of foundational studies to accurately characterize background variability, and definitive studies designed to determine the environmental impacts on germline mutation and epigenetic variation (Fig. 1). Foundational experiments must directly determine background genomic variability to identify differences between offspring and their parents in humans, and use controlled animal experiments to estimate anticipated effects sizes. These foundational studies are necessary to determine the required sample sizes and statistical approaches for pilot and definitive studies in humans. Pilot experiments should investigate highly exposed human cohorts to fully characterize dose responses. Animal models, which allow better control of genetic and environmental variables compared with humans, should be employed to strengthen causal inference in human health effects research (Table 1). Definitive studies of suspect germline mutagens must first establish biological sample repositories from human families across a broad range of potential mutagenic exposures. These studies should be prospective with well-documented epidemiological data collection, including standard epidemiological metrics, family medical history, exposure history, and samples for analyses of relevant exposure biomarkers. Definitive studies should also aim to include epigenomics, creating an integrated understanding of the epigenetic, genetic, and environmental factors explaining heritable human disease.
Fig. 1.
Recommended approach for deciphering the environmental variables that cause germline mutation and heritable epigenetic changes in humans. Establishment of baseline genetic variance in parallel with controlled experiments in animals and on highly exposed humans will provide the foundational experiments to estimate power. Tissue repositories from well-characterized human cohorts will be developed for future studies aimed at investigating the more subtle environmental variables that influence heritable outcomes. The results of these experiments will be used to develop human health policy and testing guidelines to ensure the protection of human gametes and future generations.
Table 1.
Description of the current challenges associated with applying next-generation sequencing to human families, and how inbred animals can resolve some of these issues.
| Humans | Inbred mice |
|---|---|
| High inter-individual variability will require large sample size to find an exposure effect | Decreased inter-animal variability will yield increased statistical power |
| Heterogeneous environmental exposures. | Controlled dose–response experiments can be used to derive an estimate of the environmental levels required for an effect to be observed |
| Confounding lifestyle variables | Controlled diet and environment |
| Limited tissue availability | Can sample stable non-proliferating tissues (e.g., brain); lower probability of somatic mutations |
| High error rates for sequencing technologies that are difficult to decipher from existing heterozygosities | Errors may be more readily identified in inbred animals; homozygotes |
The time is right to test the hypothesis that environmental exposures induce de novo heritable mutations in humans. This information is critical to determining the validity of extrapolations from animal models, which have already identified a number of germ-cell mutagens. These studies will determine if environmental agents that are well known to impact health in exposed individuals also cause germline mutations that affect both individual and population health in unexposed, future generations.
Acknowledgements
We thank NIH/NIEHS, NIH/NCI and Health Canada for support for this workshop and for the logistical help of the Environmental Mutagen Society, under whose auspices the workshop was organized. This manuscript was reviewed by Health Canada, the National Health and Environmental Effects Research Laboratory, the United States Environmental Protection Agency, the United States Food and Drug Administration, the National Institute of Environmental Health Sciences, National Institutes of Health, and was approved for publication. Approval does not signify that the contents reflect the views of any agency or of the United States government, nor does mention of trade names or commercial products constitute endorsement or recommendation for use.
Footnotes
The Working Committee to Develop a Protocol for Detecting Environmentally Induced Genomic Damage in the Human Germline; Hilton Montreal Bonaventure, Montreal, Quebec, Canada; October 14, 2011.
Author contributions
All authors listed are members of the ENIGMA working group, attended the ENIGMA workshop in October 2011, and participated in the round-table discussions. JBB was chair of the workshop. JJM, CLY, DMD, KLW and FM were on the organizing committee for the workshop. CLY drafted the manuscript, and all authors edited the manuscript. All authors approved the final manuscript.
Conflict of interest statement
There are no conflicts of interest for any of the authors.
References
- 1.Vissers LE, de Ligt J, Gilissen C, Janssen I, Steehouwer M, de Vries P, van Lier B, Arts P, Wieskamp N, del Rosario M, van Bon BW, Hoischen A, de Vries BB, Brunner HG, Veltman JA. A de novo paradigm for mental retardation. Nat. Genet. 2010;42:1109–1112. doi: 10.1038/ng.712. [DOI] [PubMed] [Google Scholar]
- 2.Morrow EM. Genomic copy number variation in disorders of cognitive development. J. Am. Acad. Child Adolesc. Psychiatry. 2010;49:1091–1104. doi: 10.1016/j.jaac.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lupski JR. New mutations and intellectual function. Nat. Genet. 2010;42:1036–1038. doi: 10.1038/ng1210-1036. [DOI] [PubMed] [Google Scholar]
- 4.Girirajan S, Campbell CD, Eichler EE. Human copy number variation and complex genetic disease. Ann. Rev. Genet. 2010 doi: 10.1146/annurev-genet-102209-163544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conrad DF, Pinto D, Redon R, Feuk L, Gokcumen O, Zhang Y, Aerts J, Andrews TD, Barnes C, Campbell P, Fitzgerald T, Hu M, Ihm CH, Kristiansson K, Macarthur DG, Macdonald JR, Onyiah I, Pang AW, Robson S, Stirrups K, Valsesia A, Walter K, Wei J, Tyler-Smith C, Carter NP, Lee C, Scherer SW, Hurles ME. Origins and functional impact of copy number variation in the human genome. Nature. 2010;464:704–712. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.MacArthur DG, Balasubramanian S, Frankish A, Huang N, Morris J, Walter K, Jostins L, Habegger L, Pickrell JK, Montgomery SB, Albers CA, Zhang ZD, Conrad DF, Lunter G, Zheng H, Ayub Q, DePristo MA, Banks E, Hu M, Handsaker RE, Rosenfeld JA, Fromer M, Jin M, Mu XJ, Khurana E, Ye K, Kay M, Saunders GI, Suner MM, Hunt T, Barnes IH, Amid C, Carvalho-Silva DR, Bignell AH, Snow C, Yngvadottir B, Bumpstead S, Cooper DN, Xue Y, Romero IG, Wang J, Li Y, Gibbs RA, McCarroll SA, Dermitzakis ET, Pritchard JK, Barrett JC, Harrow J, Hurles ME, Gerstein MB, Tyler-Smith C. A systematic survey of loss of function variants in human protein-coding genes. Science. 2012;335:823–828. doi: 10.1126/science.1215040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glen CD, Smith AG, Dubrova YE. Single-molecule, PCR analysis of germ line mutation induction by anticancer drugs in mice. Cancer Res. 2008;68:3630–3636. doi: 10.1158/0008-5472.CAN-08-0484. [DOI] [PubMed] [Google Scholar]
- 8.Marchetti F, Rowan-Carroll A, Williams A, Polyzos A, Berndt-Weis ML, Yauk CL. Sidestream tobacco smoke is a male germ cell mutagen. Proc. Natl. Acad. Sci. U.S.A. 2011;108:12811–12814. doi: 10.1073/pnas.1106896108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marchetti F, Wyrobek AJ. Mechanisms and consequences of paternally-transmitted chromosomal abnormalities. Birth Defects Res. C Embryo Today Rev. 2005;75:112–129. doi: 10.1002/bdrc.20040. [DOI] [PubMed] [Google Scholar]
- 10.Russell LB. Effects of male germ-cell stage on the frequency, nature, and spectrum of induced specific-locus mutations in the mouse. Genetica. 2004;122:25–36. doi: 10.1007/s10709-004-1443-7. [DOI] [PubMed] [Google Scholar]
- 11.Somers CM. Ambient air pollution exposure and damage to male gametes: human studies and in situ ‘sentinel’ animal experiments. Syst. Biol. Reprod. Med. 2011;57:63–71. doi: 10.3109/19396368.2010.500440. [DOI] [PubMed] [Google Scholar]
- 12.Witt KL, Bishop JB. Mutagenicity of anticancer drugs in mammalian germ cells. Mutat. Res. 1996;355:209–234. doi: 10.1016/0027-5107(96)00029-2. [DOI] [PubMed] [Google Scholar]
- 13.Yauk C, Polyzos A, Rowan-Carroll A, Somers CM, Godschalk RW, Van Schooten FJ, Berndt ML, Pogribny IP, Koturbash I, Williams A, Douglas GR, Kovalchuk O. Germ-line mutations, DNA damage, and global hypermethylation in mice exposed to particulate air pollution in an urban/industrial location. Proc. Natl. Acad. Sci. U.S.A. 2008;105:605–610. doi: 10.1073/pnas.0705896105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Demarini DM. Declaring the existence of human germ-cell mutagens. Environ. Mol. Mutagen. 2012;53:166–172. doi: 10.1002/em.21685. [DOI] [PubMed] [Google Scholar]
- 15.Wyrobek AJ, Mulvihill JJ, Wassom JS, Malling HV, Shelby MD, Lewis SE, Witt KL, Preston RJ, Perreault SD, Allen JW, Demarini DM, Woychik RP, Bishop JB. Assessing human germ-cell mutagenesis in the postgenome era: a celebration of the legacy of William Lawson (Bill) Russell. Environ. Mol. Mutagen. 2007;48:71–95. doi: 10.1002/em.20284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singer TM, Yauk CL. Germ cell mutagens: risk assessment challenges in the 21st century. Environ. Mol. Mutagen. 2010;51:919–928. doi: 10.1002/em.20613. [DOI] [PubMed] [Google Scholar]
- 17.Beal MA, Glenn TC, Somers CM. Whole genome sequencing for quantifying germline mutation frequency in humans and model species: cautious optimism. Mutat. Res. Rev. 2012;750(2):96–106. doi: 10.1016/j.mrrev.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 18.Roach JC, Glusman G, Smit AF, Huff CD, Hubley R, Shannon PT, Rowen L, Pant KP, Goodman N, Bamshad M, Shendure J, Drmanac R, Jorde LB, Hood L, Galas DJ. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science. 2010;328:636–639. doi: 10.1126/science.1186802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Conrad DF, Keebler JE, DePristo MA, Lindsay SJ, Zhang Y, Casals F, Idaghdour Y, Hartl CL, Torroja C, Garimella KV, Zilversmit M, Cartwright R, Rouleau GA, Daly M, Stone EA, Hurles ME, Awadalla P. Variation in genomewide mutation rates within and between human families. Nat. Genet. 2011;43:712–714. doi: 10.1038/ng.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nachman MW, Crowell SL. Estimate of the mutation rate per nucleotide in humans. Genetics. 2000;156:297–304. doi: 10.1093/genetics/156.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lynch M. Rate, molecular spectrum, and consequences of human mutation. Proc. Natl. Acad. Sci. U.S.A. 2010;107:961–968. doi: 10.1073/pnas.0912629107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lupski JR. Genomic rearrangements and sporadic disease. Nat. Genet. 2007;39:S43–S47. doi: 10.1038/ng2084. [DOI] [PubMed] [Google Scholar]
- 23.Wang J, Fan HC, Behr B, Quake SR. Genome-wide single-cell analysis of recombination activity and de novo mutation rates in human sperm. Cell. 2012;150:402–412. doi: 10.1016/j.cell.2012.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Winther JF, Boice Jr. JD, Mulvihill JJ, Stovall M, Frederiksen K, Tawn EJ, Olsen JH. Chromosomal abnormalities among offspring of childhood-cancer survivors in Denmark: a population-based study. Am. J. Hum. Genet. 2004;74:1282–1285. doi: 10.1086/421473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dubrova YE. Long-term genetic effects of radiation exposure. Mutat. Res. 2003;544:433–439. doi: 10.1016/j.mrrev.2003.05.003. [DOI] [PubMed] [Google Scholar]
- 26.Secretan B, Straif K, Baan R, Grosse Y, El Ghissassi F, Bouvard V, Benbrahim-Tallaa L, Guha N, Freeman C, Galichet L, Cogliano V. A review of human carcinogens – part E: tobacco, areca nut, alcohol, coal smoke, and salted fish. Lancet Oncol. 2009;10:1033–1034. doi: 10.1016/s1470-2045(09)70326-2. [DOI] [PubMed] [Google Scholar]
- 27.Laubenthal J, Zlobinskaya O, Poterlowicz K, Baumgartner A, Gdula MR, Fthenou E, Keramarou M, Hepworth SJ, Kleinjans JC, van Schooten FJ, Brunborg G, Godschalk RW, Schmid TE, Anderson D. Cigarette smoke-induced transgenerational alterations in genome stability in cord blood of human F1 offspring. FASEB J. 2012 doi: 10.1096/fj.11-201194. [DOI] [PubMed] [Google Scholar]
- 28.Arlt MF, Ozdemir AC, Birkeland SR, Wilson TE, Glover TW. Hydroxyurea induces de novo copy number variants in human cells. Proc. Natl. Acad. Sci. U.S.A. 2011;108:17360–17365. doi: 10.1073/pnas.1109272108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Somers CM, Cooper DN. Air pollution and mutations in the germline: are humans at risk? Hum. Genet. 2009;125:119–130. doi: 10.1007/s00439-008-0613-6. [DOI] [PubMed] [Google Scholar]
- 30.Schmitt MW, Kennedy SR, Salk JJ, Fox EJ, Hiatt JB, Loeb LA. Detection of ultra-rare mutations by next-generation sequencing. Proc. Natl. Acad. Sci. U.S.A. 2012 doi: 10.1073/pnas.1208715109. [DOI] [PMC free article] [PubMed] [Google Scholar]