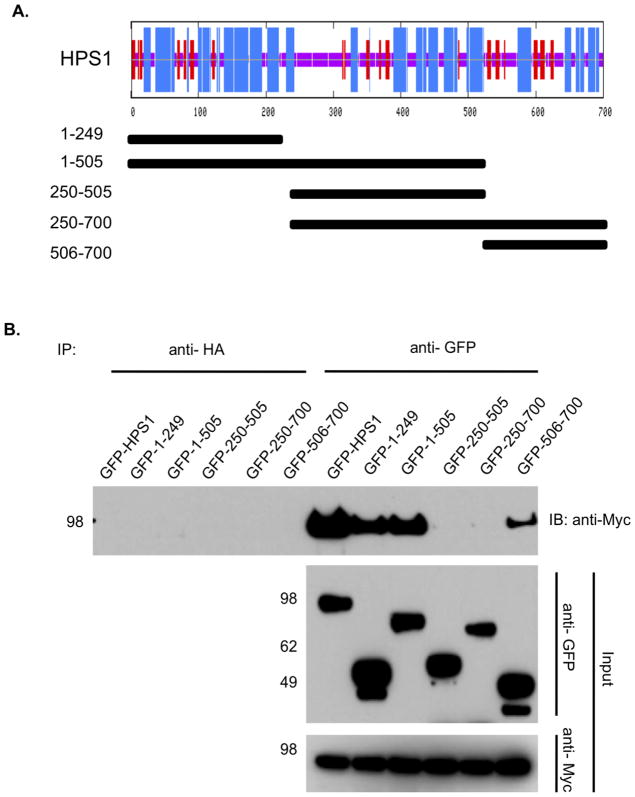
Figure 2. Divalent interaction of HPS1 with HPS4.
(A) Top. The consensus secondary structure prediction for HPS1 as determined by the Network Protein Sequence Analysis of the Pôle Bio-informatique Lyonnais. Blue areas represent α-helices, while red areas indicate predicted β-extended strand, and solid purple lines denote random coils. Bottom. GFP-tagged HPS1 constructs used to map the HPS4 binding sites on HPS1 are represented by solid bars with residue numbers indicated on the left. (B) GFP-tagged HPS1 constructs were transiently transfected into M1 clone 26 cells expressing Myc3-tagged HPS4. Coimmunoprecipitation analysis revealed an interaction of HPS4 with the first 249 amino acids of HPS1, suggesting that the N-terminus of HPS1 contains a region required for interaction with full-length HPS4. Interestingly, a construct containing the HPS1 C-terminus (HPS1506-700) also displayed an interaction with HPS4. These results are consistent with HPS4 recognizing two different regions, located at the N-terminus (first 249 amino acids) and the C-terminus (506– 700 amino acids) of HPS1. Co-immunoprecipitation with an irrelevant anti-HA antibody was used as control.