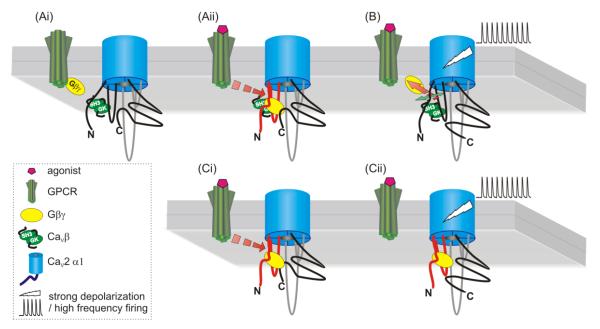
Figure 4.
Model depicting the molecular interactions that underlie Gβγ-mediated inhibition of CaV2 channels. Panels A and B (upper three images) depict a channel with a CaVβ subunit, while panel C (lower images) depicts the situation in which the CaVβ subunit is absent. Currently, data suggest the binding site for Gβγ is comprised from multiple sites on the N-terminus, I-II linker, and probably C-terminus of the channel. Binding of Gβγ causes a conformational shift that promotes interaction of the N-terminus “inhibitory module” with the initial one-third of the I-II-linker (panel Aii). This (and perhaps other interactions) shifts gating charge movement to more depolarized potentials and uncouples voltage-sensor movement from channel activation. Strong membrane depolarization (panel B) leads to conformational changes that result in unbinding of Gβγ and loss of interaction between the N-terminus and I-II linker. This depends upon binding of a CaVβ subunit to the AID on the I-II linker that induces a rigid α-helical connection to the upstream IS6 region of the pore and voltage-sensor. In the absence of CaVβ subunit binding, inhibition still occurs (panel Ci) but cannot be reversed by strong depolarization (panel Cii).