FIGURE 3. Alignment of the primary sequence of the ATases HAT core with prominent members of the GNAT superfamily.
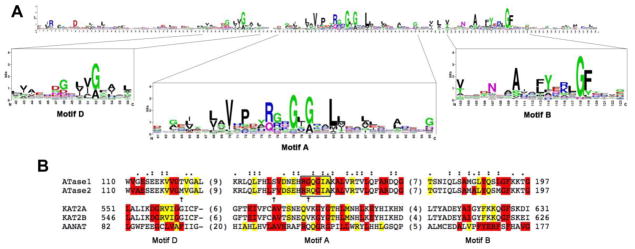
A, Sequence logo of the GNAT domain profile available at PROSITE (Database of protein domains, families and functional sites (http://prosite.expasy.org); PS51186). The sequence logo was generated after multiple sequence alignment of 660 UniProtKB/Swiss-Prot true positive hits. The consensus sequence of the GNAT domain can be read from the top of the stacks. The height of each letter is proportional to the frequency of the corresponding amino acid at the indicated position in the alignment.
B, Primary sequence alignment of the HAT core highly conserved motifs of ATase1 (NAT8B_HUMAN, Q9UHF3), ATase2 (NAT8_HUMAN, Q9UHE5) and other members of the GNAT superfamily, including Lysine acetyltransferase 2A also known as hGCN5 (KAT2A_HUMAN, Q92830), Lysine acetyltransferase 2B also known as PCAF (KAT2B_HUMAN, Q92831) and aralkylamine N-acetyltransferase (AANAT) also known as SNAT (SNAT_HUMAN, Q16613). The first and last columns indicate the amino acid position from the corresponding full sequence. The numbers in parenthesis between the conserved motifs indicate the length of the omitted sequence. The alignment was performed using the Clustal Omega multiple sequence alignment program (http://www.ebi.ac.uk/Tools/msa/clustalo). In red are highlighted fully conserved residues from the consensus sequence indicated in A. The presence of the second most frequently occurring consensus amino acid is indicated in yellow. The “:” (colon) indicates conservation between groups of strongly similar properties (scoring > 0.5 in the Gonnet PAM 250 matrix) after alignment of the ATase1 and ATase2 sequences with the GNAT consensus sequence. The “.” (period) indicates conservation between groups of weakly similar properties (scoring =< 0.5 in the Gonnet PAM 250 matrix) after alignment of the ATase1 and ATase2 sequences with the GNAT consensus sequence. The alignment of ATase1 and ATase2 sequences revealed the presence of non-conservative amino acid substitutions in motifs A and D (arrows). The box in Motif A encloses the residues that have been shown to be important for acetyl-CoA recognition and binding in other members of the GNAT superfamily.
