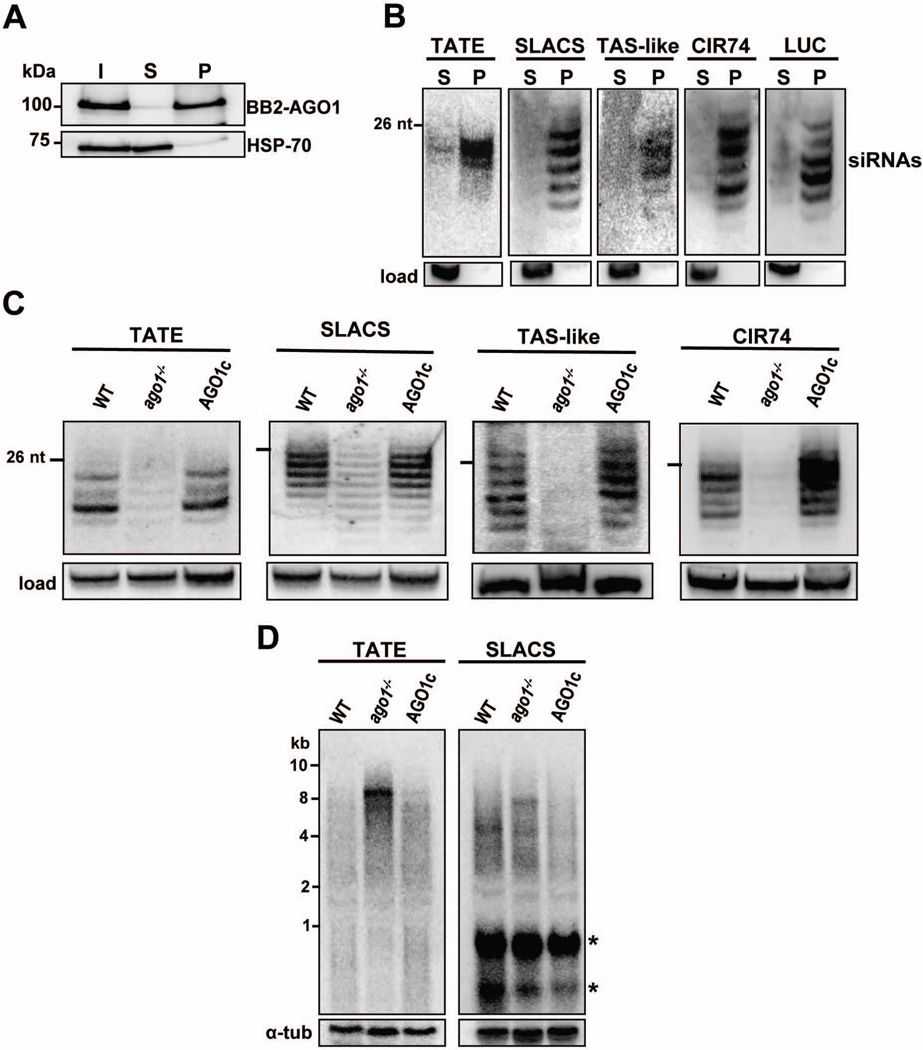
Fig. 5. LbrAGO1 controls the accumulation of siRNAs and transcripts derived from putative mobile elements.
A. Pull-down of BB2-LbrAGO1 from a soluble extract of LbrAGO1c cells. Aliquots of the input (I), supernatant (S) or the bead pellet (P), each representing 107 promastigote cells, were fractionated on a 10% SDS-PAGE and Western blotted with anti-BB2 or anti-HSP70 monoclonal antibody.
B. L. braziliensis siRNAs were extracted from immunoprecipitated BB2-LbAGO1, fractionated on a 15% sequencing gel, transferred to a nylon membrane and hybridized with the indicated probes. The membranes were stripped and re-hybridized with a 5S rRNA probe as a loading control (load). S, supernatant; P, pellet.
C. Low molecular weight RNAs from L. braziliensis wild-type (WT) promastigotes, LbrAGO1 null cells (Lbrago1−/−) or the latter complemented with the BB2-LbrAGO1 gene (LbrAGO1c) were fractionated on a 15% sequencing gel, transferred to a nylon membrane and hybridized with TATE, SLACS, TAS-like or CIR74 probes. A cross-reacting band was used as a loading control, except for the TAS-like panel where 5S RNA was the loading control.
D. Northern blot analysis of total RNA separated on a 1.2% agarose-formaldehyde gel and hybridized to a TATE or SLACS probe. The identity of the various cell lines is indicated above each lane. Hybridization to α-tubulin mRNA was used as a loading control. Asterisks denote bands cross-hybridizing with the SLACS probe.