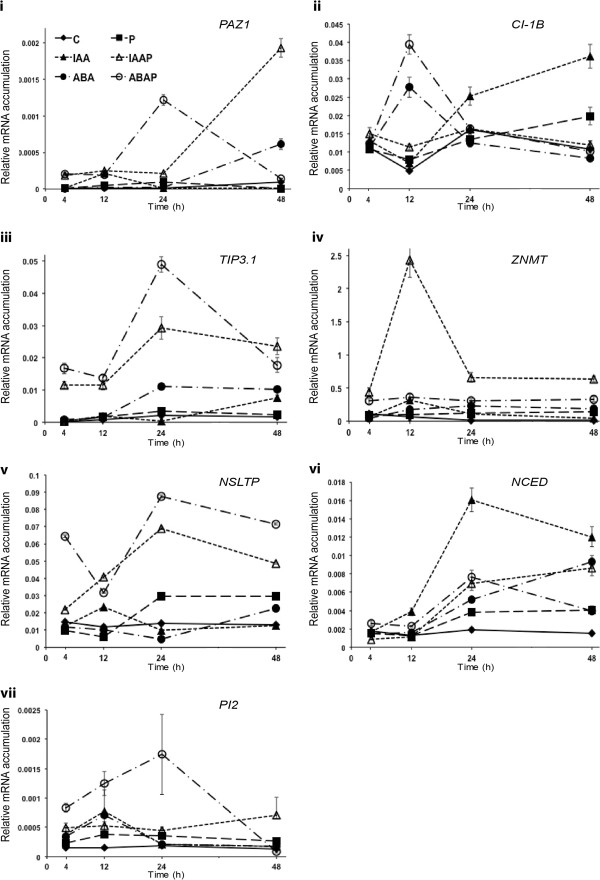
Figure 2.
Temporal analysis of the effect of hormones abscisic acid (ABA) and indole-3-acetic acid (IAA) with or without application of pathogen Fusarium culmorum on the accumulation of select transcripts in heads of barley cultivar Lux. Transcripts represented are: (a) serpin Z4 (Paz1), (b) subtilisin-chymotrypsin inhibitor (CI-1B), (c) tonoplast aquaporin (TIP3:1), (d) zinc methallothionin-like protein (ZnMT), (e) putative non-specific lipid transfer protein (nsLTP), (f) nine-cis-epoxycarotenoid dioxygenase NCED and (g) a signalling cascade protein (Pi2). Transcripts were previously identified by microarray analysis as being primed by the bacterium to accumulate in response to the pathogen at either 24 or 48 h post-pathogen treatment (a to e) or as being activated by the bacterium only (f and g). Treatments: barley heads were treated with IAA, ABA or Tween20 (control treatment), and 24 h later, with pathogen (P) or Tween20. RNA extracted from treated head tissue at either 4, 12, 24 or 48 h post-hormone or hormone and pathogen treatment was used for real-time RT-PCR analysis. aTranscript accumulation was quantified as 2-(CT target transcript–CTα-tubulin). Treatment codes: C, controls treated with Tween20; P, pathogen (F. culmorum); IAA, indole-3-acetic acid; ABA, abscisic acid; IAA + P; IAA plus pathogen; ABA+P, ABA plus pathogen. Results are based on two biological replicates, each including two techical replicates per treatment. Bars indicate standard error of mean.