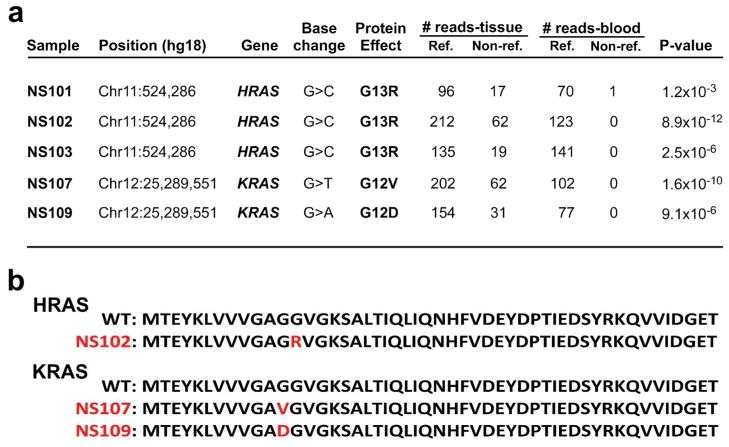
Figure 2. Exome sequencing reveals somatic HRAS and KRAS mutations in nevus sebaceus tissue.
(a) HRAS and KRAS mutation annotation, including genomic position, nucleotide change, protein consequence, and number of reference and non-reference reads obtained from paired sequencing of tissue and blood in 5 independent, unrelated nevus sebaceus cases. Significance of the mutant allele frequency difference between tissue and blood DNA was calculated with a one-tailed Fisher’s exact test. When corrected for multiple testing, 2.4×10−6 is the threshold for genome wide significance. In each case, HRAS and KRAS mutations showed the lowest P-value. (b) Alignment of the N-termini of HRAS and KRAS reveals identical residues through position 94, with an overall 95% identity and 99% similarity. The first 50 amino acids are shown for the wild-type and each mutant protein, with mutant residues indicated in red.