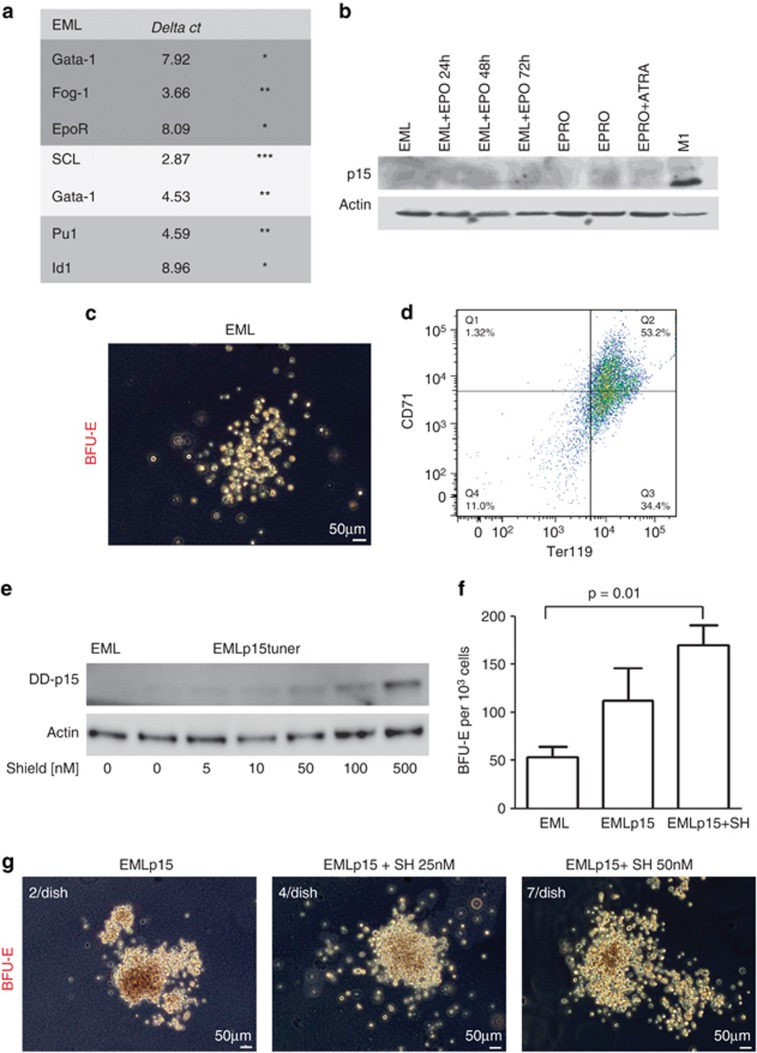
Figure 5.
Induction of p15Ink4b increases commitment of EML cells to the erythroid lineage. (a) Profiling of mRNA expression in EML cells of genes linked to erythroid (GATA-1, Fog-1, EpoR), HSC (SCL, GATA-2) or myeloid differentiation (Pu.1, Id1). Stars indicate the relative abundance of the transcript based on the ΔCt value. The Ct value inversely correlates with the gene expression. 18SrRNA was used as an internal control. (b) Lack of p15Ink4b expression in EML cells induced to differentiate towards the erythroid lineage (EML+Epo), promyelocytes (EPRO) or mature neutrophils (EPRO+ATRA). A lysate from the M1 cell line was used as a positive control. (c) Representative picture of a BFU-E colony obtained by plating 1000 EML cells in M3436 methylcellulose medium. (d) Fluorescence-activated cell sorting (FACS) analysis of the expression of Ter119 and CD71 antigens on BFU-E colonies isolated from cultured EML cells in M3436 medium for 18 days. (e) Induction of p15Ink4b in the EML cell line using the ProteoTuner system. EML cells were infected with the pLVX-PTuner-Green vector containing murine p15Ink4b cDNA cloned upstream of a destabilizing domain (DD, 12 kDa) that allowed induction of p15Ink4b expression at the protein level with minimal leakage via the addition of SH. ZSGreen-positive cells were sorted and expanded as a cell line (EMLp15Tuner, EMLp15). Expression of p15Ink4b was induced by the addition of SH to the culture medium at the indicated concentrations for 24 h. (f) Number of BFU-E colonies obtained by plating 1000 EML or EMLp15Tuner cells in M3436 methylcellulose medium pretreated with SH at a concentration of 100 nℳ for 24 h to induce the expression of p15Ink4b (n=3). (g) Representative pictures and number of large BFU-E colonies.