Abstract
The retroviral genus Lentivirus comprises retroviruses characterised from five mammalian orders. Lentiviruses typically undergo rapid rates of evolution, a feature that has allowed recent evolutionary relationships to be elucidated, but has also obscured their distant evolutionary past. However, the slowdown in the rate of evolution associated with genome invasion, as has occurred in the European rabbit, enables longer-term lentiviral evolutionary history to be inferred. Here we report the identification of orthologous RELIK proviruses in the European hare, demonstrating a minimum age of 12 million years for the lagomorph lentiviruses. This finding indicates an association between lentiviruses and their hosts covering much of the evolutionary history of the lagomorphs, and taking place within species with a worldwide distribution.
Keywords: RELIK, Hare, Lentivirus, Lepus europaeus, Evolution, Endogenous
Introduction
Lentiviruses are a genus of retroviruses that cause a range of chronic diseases in mammals, although they can be asymptomatic in some hosts. Five lentiviral subgroups are recognised by the ICTV, each restricted to a single mammalian order or family (van Regenmortel et al., 2000). Recently, a new lentiviral subgroup has been identified in the European rabbit (Oryctolagus cuniculus); RELIK (Rabbit Endogenous Lentivirus type K) is the first member of the genus to be identified in the mammalian order Lagomorpha (rabbits, hares and pikas), and is the first example of an endogenous lentivirus (i.e. a lentivirus that has entered the germline of its host) (Katzourakis et al., 2007).
Endogenous retroviruses constitute a powerful source of information for investigating the history of evolutionary interaction between viruses and their hosts. These “genomic fossils”, allow the time scale of retroviral evolution to be inferred from genomic sequence data. Most retroviruses undergo high rates of evolution, consequently events in their distant evolutionary past are difficult to reconstruct accurately using contemporary sequence data. For example, the age and ancestral host range of the primate lentiviruses remain subject to much uncertainty (Sharp et al., 2000). However, once retroviruses become endogenous, they are subjected to the background host neutral rate of substitution, which is considerably slower, facilitating the inference of evolutionary events. For example, the paired long terminal repeat (LTR) sequences generated by reverse transcription are identical at the time of integration (Telesnitsky and Goff, 1997). The time since integration can thus be estimated by measuring the divergence between these duplicated sequences and applying a molecular clock, using an appropriate calibration based on the host neutral substitution rate.
This approach can be extended to any pairs of duplicated sequences that have been evolving neutrally, e.g. pairs of ERV sequences generated by segmental duplication events in the host genome. As substitution rates in host genomes can be several orders of magnitude slower than in replicating retrovirus populations (Holmes, 2003; Kumar and Subramanian, 2002), and relatively free of rapidly changing selective pressures, molecular clock estimations that extend millions of years backward in time are more reliable for ERVs than for exogenous viruses. Analysis of segmentally duplicated insertions led to an estimated minimum age of ~7–11 million years for the RELIK subgroup, establishing that the evolution of lentiviruses should be considered on a time scale spanning several millions of years, rather than the shorter time frames suggested by analysis of exogenous sequence data (Holmes, 2003; Katzourakis et al., 2007).
The reliability of estimates based on divergence between pairs of sequences is determined by the accuracy of the calibration rate used. An alternative and more robust approach is the identification of orthologous ERV insertions in multiple species. The identification of such orthologues indicates that integration must have occurred prior to the divergence of the respective hosts, and can thus provide an estimated minimum age based on the estimated time since host divergence. This estimated minimum will always be an underestimate of the age of the provirus, as the shared insertion must predate the speciation event of the host species, and could do so by thousands or even millions of years. In common with approaches relying on duplicated sequences, the reliability of these estimates will be dependent on the accuracy of the molecular clock that is used to estimate the time of divergence of the hosts in which the orthologues are found. These divergence dates can combine multiple host genes, as well as paleontological data, and can also relax the assumption of a strict molecular clock (Drummond et al., 2006; Matthee et al., 2004). Analyses based on orthologous integrations have been used to provide robust support for phylogenetic relationships between species, and to discriminate between otherwise contentious alternative evolutionary hypotheses (Johnson and Coffin, 1999; Lunter, 2007). Here, we report the identification of orthologous lentiviral insertions in the European rabbit and the European brown hare (Lepus europaeus), and examine the implications for lentiviral evolution.
Results and discussion
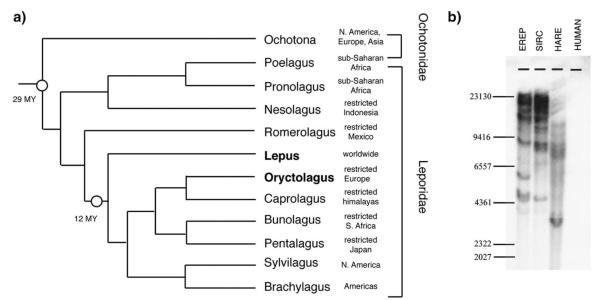
Phylogenetic relationships in the order Lagomorpha are illustrated in Fig. 1. The order includes two families, Ochotonidae (pikas) and Leporidae (hares and rabbits), and more than 80 species (Angerman et al., 1990; Matthee et al., 2004; Yu et al., 2000). The family Ochotonidae contains 26 species (Yu et al., 2000), one of which, the American pika (Ochotna princeps), is the subject of an ongoing genome sequencing project. Screening of the currently available 7 × 109 nucleotides of the low coverage (2x) O. princeps genome using sequence probes derived from lentiviral coding regions and LTRs failed to identify any significant matches. This observation, coupled with the relatively ancient date of the Ochotonidae–Leporidae divergence (~29 mya (Matthee et al., 2004)) relative to the oldest estimated dates for RELIK insertions (~11 mya), makes it unlikely that RELIK insertions orthologous to those identified in the O. cuniculus genome are present in O. princeps or other species in the family Ochotonidae. However, as the oldest estimated dates for RELIK insertions represent a conservative lower estimate for germ-line infection with RELIK, this infection could have occurred at any time between the Ochtonidae–Leporidae divergence and this date. The unoccupied preintegration site could not be detected in the genome assembly of O. princeps using BLAST searches, however the multi-copy nature of the RELIK subgroup renders it unlikely that copies are present in the pika genome.
Fig. 1.
(a) Lagomorph evolutionary timeline, adapted from (Matthee et al., 2004). Geographic distributions are shown for each genus. The 95% confidence intervals for the node ages are 22.44–37.15 mya for the most basal node indicated, and 9.79–14.54 for the node below the Lepus branch (Matthee et al., 2004). (b) Southern probing for a RELIK gag–pol fragment. SIRC and EREP are rabbit cell lines. Hare DNA was extracted from a hare ear. Human DNA is from HeLa cells. 20 μg of each was cut with HindIII and run on the gel, transferred to a nylon membrane and probed with a P32 dCTP labelled fragment of RELIK. Southern blot is representative of two experiments performed with independently prepared DNA samples. Size markers (Lambda HindIII) are shown in base pairs.
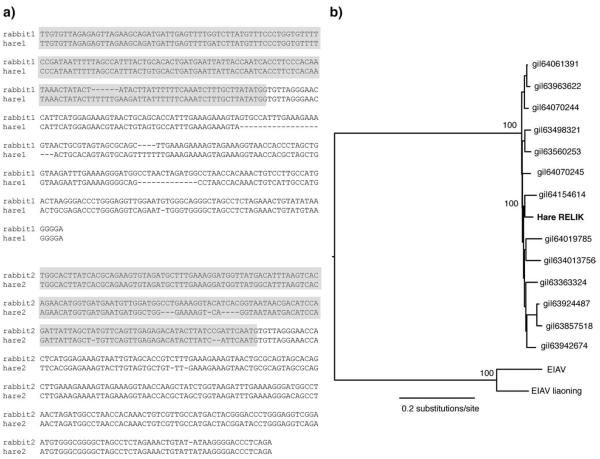
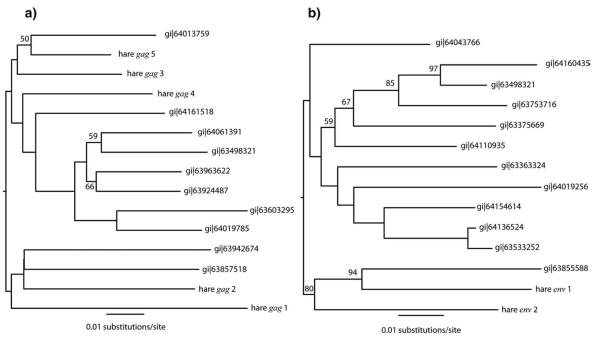
We sought evidence for the presence of RELIK insertions in L. europaeus by Southern blot (Fig. 1b). We probed Hind3 cut genomic DNA from 2 rabbit cell lines (SIRC, EREP) and a European brown hare with radioactively labelled DNA derived by PCR from rabbit genomic DNA. The Southern blot revealed the presence of multiple insertions in the rabbit genome and at least 5 insertions in the hare genome. Next, we sought RELIK orthologues in the L. europaeus genome by PCR, employing a screening strategy that utilized primer pairs directed against genomic sequences spanning both a RELIK LTR and the adjacent region of host DNA flanking it (see Fig. 2a). Primers were designed using O. cuniculus sequences identified in a BLAST search of whole genome shotgun (WGS) assembled contigs using a single RELIK LTR as a probe. This identified a total of 27 contigs that included a minimum 100 nt of intact 5′ or 3′ RELIK LTR and 200 nucleotides of adjacent flanking sequence. To exclude insertions in highly repetitive regions (e.g. inserted into another transposable element such as a SINE), each flanking region was used as a probe in a separate BLAST search of the O. cuniculus genome. Flanking sequences that returned >10 near matches were considered likely to constitute repetitive DNA, and were not targeted for PCR amplification. PCR screening of L. europaeus genomic DNA using 11 pairs of primers targeted against RELIK LTRs and non-repetitive flanking regions generated two distinct amplicons (Fig. 2b). To determine whether the hare RELIK insertions fell within the diversity of rabbit RELIK insertions, we also amplified the pol gene. Maximum likelihood (ML) phylogenetic trees based on the amplified fragment aligned against an existing multiple alignment of RELIK sequences, and including EIAV sequences as an outgroup, confirmed that the sequence from the European hare fell within the existing RELIK subgroup diversity (Fig. 2b). Furthermore, additional phylogenetic trees obtained from the gag and pol genes, identified multiple phylogenetically interspersed RELIK insertions in the hare genome (Fig. 3).
Fig. 2.
(a) Orthologous RELIK inserts, spanning the flanking region and 5′ LTR in the hare and rabbit genomes. The region of the sequence boxed in grey indicates the flanking region. Rabbit1 corresponds to the RELIK sequence found in AAGW01527435 while rabbit2 corresponds to the RELIK sequence found in AAGW01579769. (b) ML phylogeny of hare RELIK pol sequence together with rabbit RELIK sequences, rooted using EIAV outgroup sequences. For clarity, only the bootstraps (1000 replicates) for the whole RELIK clade and for the hare RELIK sequence against the most closely related rabbit sequence are shown.
Fig. 3.
(a) ML phylogeny of 5 gag gene fragments from hare RELIK insertions, together with rabbit RELIK sequences. Numbers represent bootstrap percentages (1000 replicates). (b) ML phylogeny of 2 env gene fragments from hare RELIK insertions, together with rabbit RELIK sequences. Bootstrap values displayed as in (a).
The identification of RELIK orthologues in the hare indicates that integration occurred prior to the divergence of the Lepus and Oryctolagus genera ~12 million years ago (Matthee et al., 2004), and therefore implies the presence of RELIK insertions in the entire Lepus genus. The monotypic genus Oryctolagus, in which the RELIK subgroup was first identified, has a relatively restricted geographical distribution. By contrast, the Lepus genus (hares and jackrabbits) has ~26 species and a cosmopolitan distribution (Flux and Angerman, 1990). Furthermore, the presence of RELIK insertions in the Lepus/Oryctolagus ancestor, and the paraphyletic nature of this divergence (Fig. 1), implies the presence of RELIK insertions in all the species that arose subsequent to this node. This would include the monotypic genera Bunolagus, Caprolagus, Pentalagus and Brachylagus, all of which have relatively restricted, but non-overlapping distributions (African, Asian, South East Asian and North American, respectively). Additionally, RELIK may be present in Sylvilagus, a genus containing more than 16 species, with a wider distribution, found in North, Central, and South America (Chapman and Ceballos, 1990). Thus, potentially, RELIK-like sequences may be present in 11 genera of Leporidae, comprising more than 50 species in total, with a worldwide distribution. The implied host distributions of RELIK insertions rely on the assumption that integration occurred prior to speciation of the hosts. If integration occurred close to the time of speciation such that integration sites were still polymorphic in the host population, it is possible that some descendant lineages inherited chromosomes without the integration.
It is possible that RELIK may have gone extinct via recombinational deletion in some of these species (Katzourakis et al., 2005). Previous analysis of RELIK in O. cuniculus showed that all copies in the rabbit genome were defective. However, ERVs can retain replication competence for many millions of years (Belshaw et al., 2004; Patience et al., 1997). The ancestral insertions identified here indicate that RELIK must have been infecting the germline of the ancestor of the Lepus and Oryctolagus genera, and it remains possible that active circulating RELIK-like viruses are present in some species of Leporidae at the present day. It is not currently possible to ascertain the extent to which RELIK circulated as an exogenous lentivirus in lagomorphs, occasionally infecting the germline, rather than having infected the germline on a single occasion >12 mya followed by proliferation within the genome. We note that rabbits possess an active TRIM5 protein that exhibits strong restriction against a range of retroviruses, and it is likely that this restriction system is derived from an antiviral ancestral gene that is common to mammals (Schaller et al., 2007; Ylinen et al., 2006). The age, and inferred widespread nature among lagomorphs of RELIK could have provided, at least in part, a strong selection pressure for the maintenance of rabbit TRIM5 activity.
Materials and methods
Southern hybridisation and PCR amplification
We blotted Hind3 cut genomic DNA from 2 rabbit cell lines (SIRC, EREP), 1 human cell line (HeLa) and the ear of a European brown hare (Qiaprep, Qiagen UK). The hare sample was obtained from an exotic meats butcher (McKenna meats). We probed the blot with radioactively labelled (Rediprime, GE Healthcare) RELIK pol DNA which was PCR amplified from the rabbit cell line using primers forward REL9 5′-GCAATGCCCCCCGGACCATGATGGC-3′ and reverse REL2rev 5′-ATGGCTACCAGAATTAGCCGGGCCTCATAGTG-3′. The blot was washed at high stringency (0.1 × SSC, 0.1% SDS) and revealed by exposure to film. Ultraviolet examination of the agarose gel after staining with ethidium bromide but before transfer to PVDF indicated that the tracks were loaded with similar amounts of DNA (results not shown).
Hare flanking sequence 1 was PCR amplified using platinum pfx (Invitrogen) according to manufacturer’s instructions and primers forward GT464 5′-GTGTTAGAGAGTTAGAAGCAG-3′ and reverse REL13 5′-CCCCTTATATACAGTTTCTAGAGGC-3′. For Hare flanking sequence 2 primers were forward GT469 5′-GGCACTTATCACGCAGAAGTG-3′ and reverse GT460 5′-GTTTACAGCGTCTGAGGGTCCC-3′. Hare Pol sequence was amplified using primers REL9 and REL2rev as described above. Hare Gag sequence was amplified using the primers forward REL1fwd 5′-TGTTAGGGAACCATTCACAGAGAAAGTAATTG-3′ and reverse Hare seq + 2045 5′-CCCCCTAGGTTTACCTTTAAGGTAGG-3′. Hare Env sequence was amplified using the primers forward REL5fwd 5′-ACCTTTGAACAAAACAGGGGAGTCCAAATAGGGTAGGGACAAGAAAAG-3′ and reverse REL5rev 5′-AAGCATACAAGAACCATACAAAATATTGCTCC-3′. PCR amplifications were repeated on DNA extracted from a kidney cell line derived from a European brown hare (a gift from Jean Francois Vautherot).
Phylogenetic analysis
The amplified pol gene from the hare was aligned against an existing multiple alignment of rabbit RELIK sequences (Katzourakis et al., 2007). ML phylogenetic trees were constructed under a successive approximations heuristic searching strategy in PAUP* (Swofford, 2003), using an initial neighbour joining tree, followed by two rounds of branch swapping (TBR and NNI), with parameter optimisation between each round. EIAV (NC_001450) and EIAV liaoning (AF327877) were used as outgroups for phylogenetic analysis of pol. Support was evaluated with 1000 neighbour-joining bootstrap replicates, using the maximum likelihood distances estimated for the ML tree. ML phylogenetic trees for the gag and env genes were estimated, and support evaluated, under the same procedure. Due to the lack of an outgroup with a sufficiently long alignable region, the phylogenetic trees were mid-point rooted.
Nucleotide sequences and accession numbers
The hare RELIK sequences generated in this study have been submitted to GenBank under accession numbers FJ493029, FJ493030, FJ493031, FJ493032, FJ493033, FJ493034, FJ493035, FJ493036, FJ493037 and FJ493038.
Acknowledgments
We would like to thank M. Tristem and Bodo Schulenburg for helpful suggestions, Jurgen Roes and Marieke Bokhoven for advice and reagents, and Jean Francois Vautherot for the gift of the hare kidney cell line. ZK was funded by the UCL graduate school. GJT and LMJY were funded by Wellcome Trust Senior Fellowship No 076608 to GJT. AK was funded by the MRC and the James Martin 21st Century School.
References
- Angerman R, Flux JEC, Chapman JA, Smit AT. Lagomorph classification. In: Chapman JA, Flux JEC, editors. Rabbits, hares and pikas: status conservation action plan. International union for conservation of nature and natural resources; Gland, Switzerland: 1990. pp. 7–13. [Google Scholar]
- Belshaw R, Pereira V, Katzourakis A, Talbot G, Paces J, Burt A, Tristem M. Long-term reinfection of the human genome by endogenous retroviruses. Proc. Natl. Acad. Sci. U.S.A. 2004;101:4894–4899. doi: 10.1073/pnas.0307800101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman JA, Ceballos G. The cottontails. In: Chapman JA, Flux JEC, editors. Rabbits, hares and pikas: Status conservation action plan. International Union for Conservation of Nature and Natural Resources; Gland, Switzerland: 1990. pp. 95–110. [Google Scholar]
- Drummond AJ, Ho SYW, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. Plos Biology. 2006;4:699–710. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flux JEC, Angerman R. The hares and jackrabbits. In: Chapman JA, Flux JEC, editors. Rabbits, hares and pikas: Status conservation action plan. International Union for Conservation of Nature and Natural Resources; Gland, Switzerland: 1990. pp. 61–94. [Google Scholar]
- Holmes EC. Molecular clocks and the puzzle of RNA virus origins. J. Virol. 2003;77(7):3893–3897. doi: 10.1128/JVI.77.7.3893-3897.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson WE, Coffin JM. Constructing primate phylogenies from ancient retrovirus sequences. Proc. Natl. Acad. Sci. U. S. A. 1999;96(18):10254–10260. doi: 10.1073/pnas.96.18.10254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzourakis A, Rambaut A, Pybus OG. The evolutionary dynamics of endogenous retroviruses. Trends Microbiol. 2005;13(10):463–468. doi: 10.1016/j.tim.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Katzourakis A, Tristem M, Pybus OG, Gifford RJ. Discovery and analysis of the first endogenous lentivirus. Proc. Natl. Acad. Sci. U. S. A. 2007;104(15):6261–6265. doi: 10.1073/pnas.0700471104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Subramanian S. Mutation rates in mammalian genomes. Proc. Natl. Acad. Sci. U. S. A. 2002;99(2):803–808. doi: 10.1073/pnas.022629899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunter G. Dog as an outgroup to human and mouse. Plos Comput. Biol. 2007;3(4):772–774. doi: 10.1371/journal.pcbi.0030074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthee CA, van Vuuren BJ, Bell D, Robinson TJ. A molecular supermatrix of the rabbits and hares (Leporidae) allows for the identification of five intercontinental exchanges during the Miocene. Syst. Biol. 2004;53(3):433–447. doi: 10.1080/10635150490445715. [DOI] [PubMed] [Google Scholar]
- Patience C, Takeuchi Y, Weiss RA. Infection of human cells by an endogenous retrovirus of pigs. Nat. Med. 1997;3(3):282–286. doi: 10.1038/nm0397-282. [DOI] [PubMed] [Google Scholar]
- Schaller T, Hue S, Towers GJ. An active TRIM5 protein in rabbits indicates a common antiviral ancestor for mammalian TRIM5 proteins. J. Virol. 2007;81(21):11713–11721. doi: 10.1128/JVI.01468-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PM, Bailes E, Gao F, Beer BE, Hirsch VM, Hahn BH. Origins and evolution of AIDS viruses: estimating the time-scale. Biochem. Soc. Trans. 2000;28(2):275–282. doi: 10.1042/bst0280275. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP*: phylogenetic analysis using parsimony (* and other methods) 4 ed Sinauer Associates; Sunderland, MA: 2003. [Google Scholar]
- Telesnitsky A, Goff SP. Reverse transcriptase and the generation of retroviral DNA. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. Cold Spring Harbor Laboratory Press; New York: 1997. pp. 121–160. [PubMed] [Google Scholar]
- van Regenmortel MHV, Fauquet CM, Bishop DHL, Carstens EB, Estes MK, Lemon SM, Maniloff J, Mayo MA, McGeoch DJ, Pringle CR, Wickner RB. Virus Taxonomy: The Classification and Nomenclature of Viruses. The Seventh Report of the International Committee on Taxonomy of Viruses. Academic Press; San Diego: 2000. [Google Scholar]
- Ylinen LMJ, Keckesova Z, Webb BLJ, Gifford RJM, Smith TPL, Towers GJ. Isolation of an active Lv1 gene from cattle indicates that tripartite motif protein-mediated innate immunity to retroviral infection is widespread among mammals. J. Virol. 2006;80(15):7332–7338. doi: 10.1128/JVI.00516-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu N, Zheng CL, Zhang YP, Li WH. Molecular systematics of pikas (genus Ochotona) inferred from mitochondrial DNA sequences. Mol. Phylogenet. Evol. 2000;16(1):85–95. doi: 10.1006/mpev.2000.0776. [DOI] [PubMed] [Google Scholar]