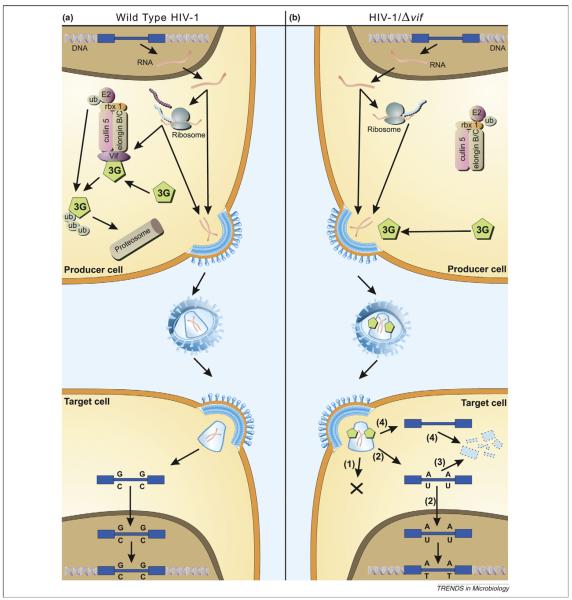
Figure 1.
Model of APOBEC antiviral function. (a) Effect of hA3G on the life cycle of wild-type HIV-1. The integrated HIV-1 provirus (blue) is transcribed and translated like a normal cellular gene. The viral Vif protein (purple) acts as an adaptor protein, connecting hA3G (green) to an E3 ubiquitin ligase complex comprising ElonginB, ElonginC, Cullin5 and RING-box-1 (rbx1), thereby inducing the polyubiquitylation and proteasomal degradation of hA3G. Viral proteins (turquoise) and genomic RNA (red) assemble and bud through the plasma membrane, releasing viral particles. Virions then enter target cells and, after reverse transcription, uncoating and nuclear entry have taken place, the viral DNA is integrated into the host genome. Formation of this provirus thus completes the replication cycle. (b) Effect of hA3G on the life cycle of HIV-1/Δvif. The HIV-1 provirus is transcribed and translated as before, but no Vif is synthesized. Cellular hA3G is therefore not degraded but instead is packaged into nascent budding virions. On infection of target cells, the presence of hA3G leads to a block in infection by any one or more of the following mechanisms. (1) hA3G might impede the formation of reverse transcripts through an unknown mechanism. (2) Cytidine deamination of nascent reverse transcripts by hA3G could be sufficient to hinder ensuing rounds of infectious virion production owing to inactivating mutations in viral genes and/or proteins. (3) Edited reverse transcripts might be recognized by cellular DNA repair pathways, leading to the degradation of these transcripts. (4) It is also possible that hA3G induces the degradation of HIV-1 reverse transcripts through an editing-independent mechanism, perhaps by recruiting a cellular endonuclease. (Reproduced with permission from [6]).