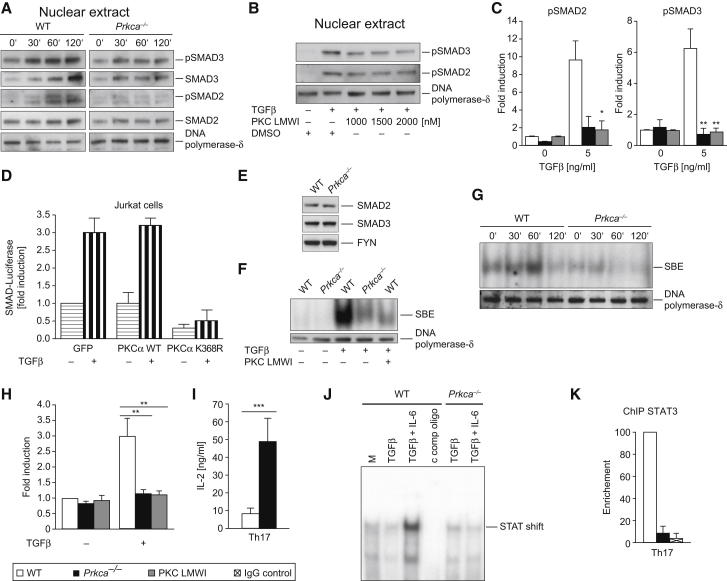
Figure 5.
The Activation of SMAD2 and SMAD3 by TGFβRI Critically Depends on PKCα
(A and B) IB shows that TGFβ stimulation leads to SMAD2 and SMAD3 phosphorylation in WT cells yet is hindered in both Prkca−/− cells (A) and cells preincubated with PKC LMWI (low-molecular-weight inhibitor) (B). CD4+ T cells were treated with TGFβ as indicated, and nuclear extracts (NEs) were generated.
(C) Graphs represent the fold induction of normalized nuclear levels of selected TGFβ concentrations (0 and 5 ng/ml) of (p)SMAD2 (left) and (p)SMAD3 (right).
(D) Transfection of the catalytically inactive kinase-dead K368R PKCα mutant (but not the WT) expression vector in Jurkat T cells repressed the expression of a SMAD2-3-dependent promoter luciferase reporter.
(E) IB analysis confirmed equal total-protein expression levels of SMAD2 and SMAD3 between WT and Prkca−/− CD4+ T cell lysates.
(F and G) Reduced SMAD2-3 DNA binding to the SMAD-binding element (SBE) in both Prkca−/− cells and PKC-LMWI-preincubated cells, as determined by electromobility shift assays (EMSA). CD4+ T cells were treated with TGFβ as indicated, and NEs were generated.
(H) The graph represents the fold induction of normalized SMAD2-3-DNA-binding efficiency.
(I) IL-2 levels were analyzed in Th17-cell-differentiated WT and Prkca−/− cells. Data are from four independent experiments.
(J) EMSA analysis of the binding capability of an Il17a minimal promoter oligonucleotide, containing a consensus STAT binding site and incubated with WT or Prkca−/− NEs of cells that were stimulated as indicated (2 hr). Resting cells were used as unspecific controls.
(K) Chromatin immunoprecipitation (ChIP) analysis (for 16 hr) of the binding capability of STAT3 to the minimal Il17a promoter in Th17-cell (TGFβ + IL-6)-primed, WT, or Prkca−/− CD4+ T cells. Data are from two independent experiments.
Representative blots are from two (J) or at least three (A, B, and E–G) independent experiments that yielded similar results. In (C) and (H), the results are presented relative to unstimulated WT levels and are normalized to the expression levels of DNA polymerase-δ, and data were pooled from at least three independent experiments. Error bars in (C), (D), (H), (I), and (K) represent the mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. (See also Figure S5).