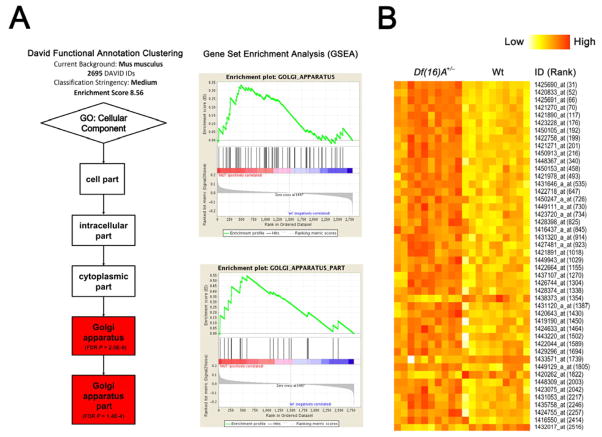
Figure 5. Coordinated mild dysregulation of Golgi-related putative miR-185 targets in Df(16)A+/− mice.
(A) DAVID functional annotation clustering analysis (left) and Gene Set Enrichment Analysis (GSEA v2.0) (right) of genes predicted as miR-185 targets by TargetScan Mouse v5.2 identified Gene Ontology (GO) terms “Golgi apparatus” and “Golgi apparatus part” as the top enriched gene sets (see Supplemental Methods).
(B) Expression heatmap plot of the potential miR-185 targets that serve Golgi apparatus related functions (GO term) and are differentially expressed (P < 0.005) between adult HPC of Df(16)A+/− mice and Wt littermates. ID is Affymetrix ID (see Table S4); Rank is the ranking position in the list of all differentially expressed genes according to significance level. Note that the majority (89%, 34 out of 38) of the genes are upregulated. See also Figure S5 and Table S4.