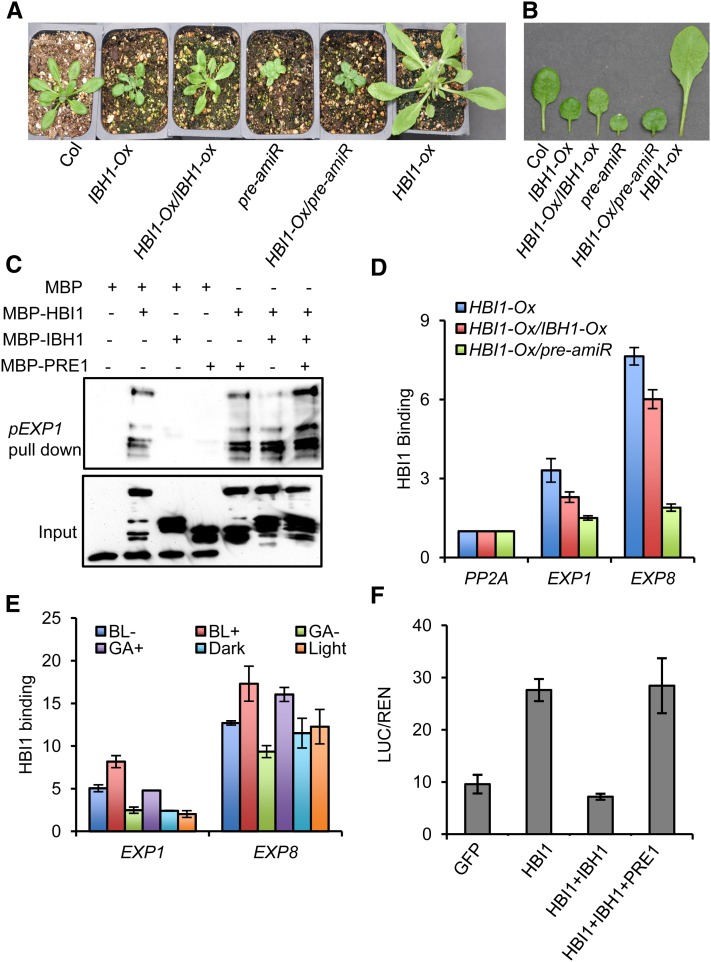
Figure 6.
IBH1 and PRE1 Antagonistically Regulate the Activity of HBI1.
(A) and (B) Overexpression of IBH1 or knockdown the expression of PREs suppresses the phenotype of HBI1-Ox. Plants (A) or detached leaves (B) were photographed after growth in soil for 4 weeks.
(C) In vitro DNA pull-down assays of HBI1 DNA binding activity. The indicated MBP or MBP fusion proteins purified from Escherichia coli were incubated with a biotinylated DNA fragment of the EXP1 promoter immobilized on streptavidin beads. The DNA-bound proteins were immunoblotted using anti-MBP antibody.
(D) ChIP-qPCR analysis of HBI1 binding to the EXP1 and EXP8 promoters and the effects of IBH1-Ox and pre-amiR on HBI1 DNA binding in vivo. Heterozygous transgenic 35S:HBI1-YFP/Col-0, 35S:HBI1-YFP/35S:IBH1-myc, and 35S:HBI1-YFP/35S:pre-amiR F1 plants grown in a greenhouse for 4 weeks were used for the ChIP-qPCR analysis. Error bars indicate sd of three biological repeats.
(E) ChIP-qPCR analysis of the effects of BR, GA, and light on HBI1 DNA binding in vivo. ChIP-qPCR was performed using 35S:HBI1-YFP and 35S:YFP grown in half-strength MS liquid medium with or without 2 µM PPZ or 1 µM PAC for 5 d under constant light or in the dark. The plants grown on 2 µM PPZ were treated with mock solution (BL−) or 100 nM BL (BL+) for 6 h; plants grown on 1 µM PAC were treated with mock solution (GA−) or 10 µM GA3 (GA+) for 6 h; plants grown in the dark were treated with white light (35 µM/m2/s) or kept in the dark for 6 h. Error bars indicate sd of three biological repeats.
(F) Transient assays show HBI1 activation of the pEXP1:LUC reporter gene. Arabidopsis protoplasts were transformed with the dual luciferase reporter construct containing pEXP1:LUC (luciferase) and 35S:REN (renilla luciferase) and constructs overexpressing the indicated effecters. The LUC activity was normalized to REN. Error bars indicate sd of three biological repeats.