Fig. 1.
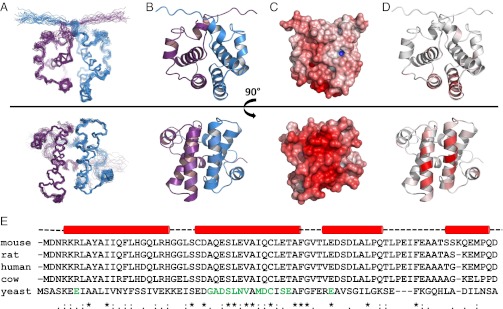
NMR structures of Sgt2_NT dimer rotated 90° around the x-axis. (A) Ensemble views with monomers represented in blue and violet. (B) Ribbon representation with monomers represented in blue and violet. (C) Electrostatic views ranging from –10 negative charge in red to +10 positive charge in blue modeled using the adaptive Poisson-Bolzmann solver (APBS) PyMol plug-in, which calculates the charge distribution displayed on the solvent accessible surface of the protein. (D) Ribbon views colored according to chemical shift perturbation upon binding to Get5_UBL. Residues greater than 80% of maximum chemical shift are colored the darkest red. Between 0% and 80% is divided equally across the seven remaining shades. (E) Sequence alignment of SGT proteins from mouse, rat, human, cow, and yeast with sequence conservation and similarity indicated below. Residues known, from this study, to participate in binding to UBL domains are indicated in green. Helical secondary structure above is derived from our structure solution in yeast. The binding surface is predominantly localized to the second helix (in both yeast and human according to results in this article) and hence the helical dimer interface.
