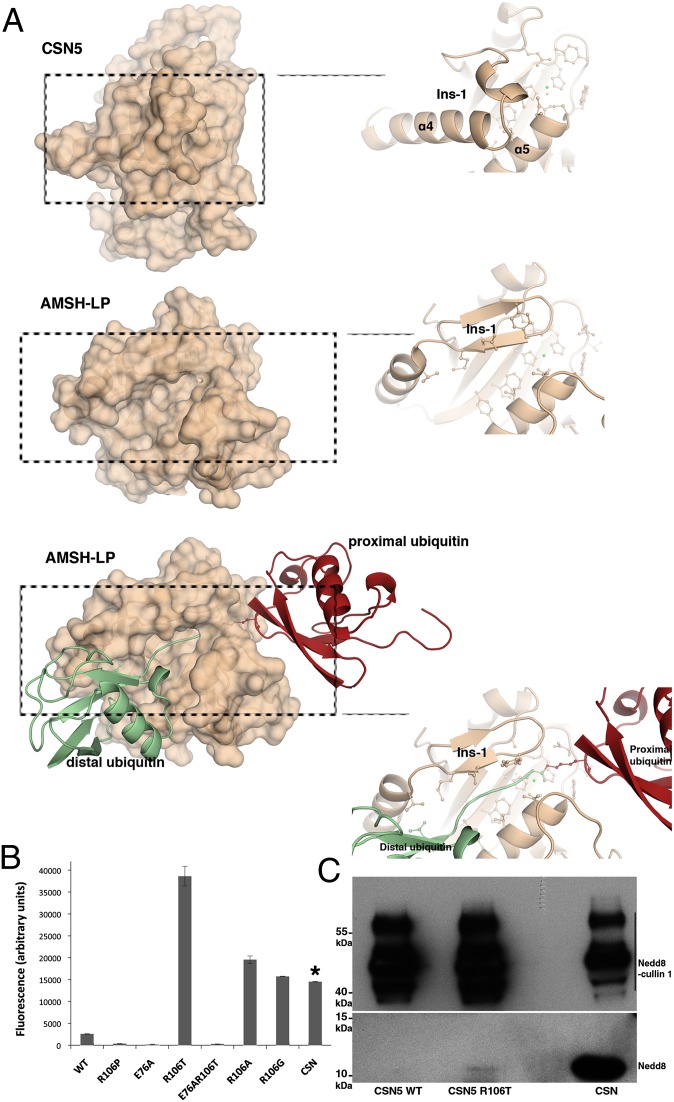
Fig. 4.
CSN5 adopts a conformation incompatible with Nedd8 recruitment and requires conformational relaxation to perform catalysis. (A) CSN5 Ins-1 masks the putative binding site for Nedd8. (Top) CSN5 surface and a close-up view of the Ins-1 region. (Middle) AMSH-LP surface and a close-up view of the Ins-1 region. (Bottom) Surface of AMSH-LP bound to K63-Ub2 and a close-up view of the groove that accommodates the isopeptide. (B) R106T (or R106A,G) substitution constitutively activates CSN5 against Nedd8 substrate. Isopeptidase activity of CSN5 or of the CSN on Nedd8-AMC was measured by the increase of fluorescence intensity at 460 nm (λexcitation = 380 nm) after 50 min at 28 °C. The concentration of CSN5 used in these experiments is 210-fold more than that of the CSN complex, as indicated by an asterisk. Taking into account this dilution factor, the CSN complex appears 79-fold more active than the most active R106 variant, R106T. (C) The R106T variant form of CSN51–257 is able to deneddylate Nedd8-cullin 1. (Upper) Nedd8 signal of the Nedd8-cullin 1 and (Lower) Nedd8 released on Nedd8-cullin 1 isopeptide bond hydrolysis for CSN5 WT, CSN5 R106T, and the CSN complex (from left to right).