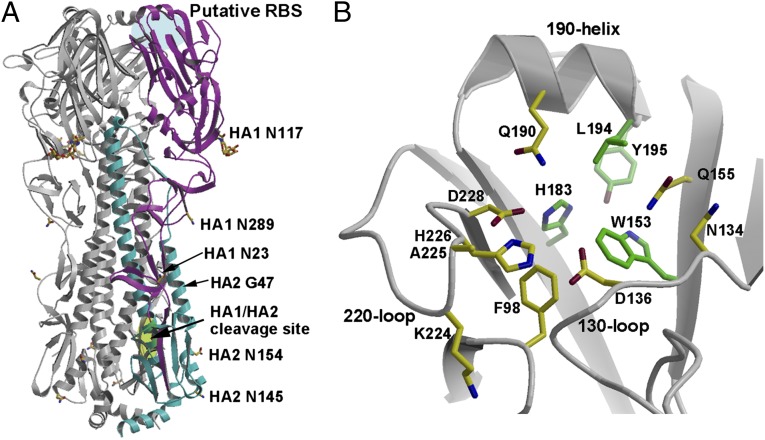
Fig. 1.
Crystal structure of GU10-060 HA2-47G HA. (A) Overview of the H17 HA trimer. For clarity, one of the monomers is highlighted in magenta (HA1) and cyan (HA2). The receptor-binding site (RBS) in influenza A and B HAs is highlighted in blue and designated here as the HA putative RBS because no binding activity has yet been found for H17 HA. The HA1/HA2 single Arg cleavage site is highlighted in green. Carbohydrate observed at HA1 Asn117 in the electron density map is colored yellow, and other asparagines, 23 and 289 of HA1, and 145 and 154 of HA2, that code for potential N-glycosylation sites are also labeled. The GU10-060 HA2 A47G mutation was made to stabilize an ectodomain trimer during baculovirus expression; the HA2 47 position is on the trimer interface and far from the RBS. (B) Putative RBS of the H17 HA with key side chains shown in sticks. The four residues that are conserved in the H17 HA and other HAs are colored with green carbon atoms, whereas other nonconserved putative RBS residues are colored with yellow carbon atoms.