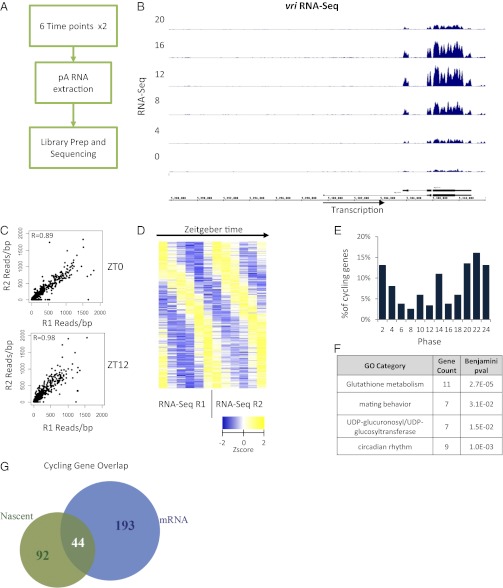
Fig. 2.
RNA-Seq identifies more than 237 mRNA cyclers in Drosophila heads. (A) Overview of the workflow used to prepare and sequence rhythmic mRNA. (B) Cycling mRNA signal over the vri locus for one set of replicates. Signal within the intron is depleted. (C) The reads per base pair coverage (blue) was used to quantify the average coverage within exons. The correlation of this measure between the replicate time points is good, with coefficients of R = 0.89 and R = 0.98 for ZT0 and ZT12, respectively. (D) Fourier analysis of RNA-Seq datasets identifies 237 mRNA cyclers. Shown are the Z-scores of the reads per base pair coverage plotted as a heatmap. (E) The phase distribution of cycling is more spread out than the Nascent-Seq cyclers and increases gradually, peaking at ZT22. (F) A few GO categories were enriched, including circadian rhythms and glutathione metabolism. (G) A significant overlap between the Nascent and mRNA cycling genes is observed. Forty-four genes have robust Nascent and mRNA cycling which is indicative of the classic circadian transcription model.