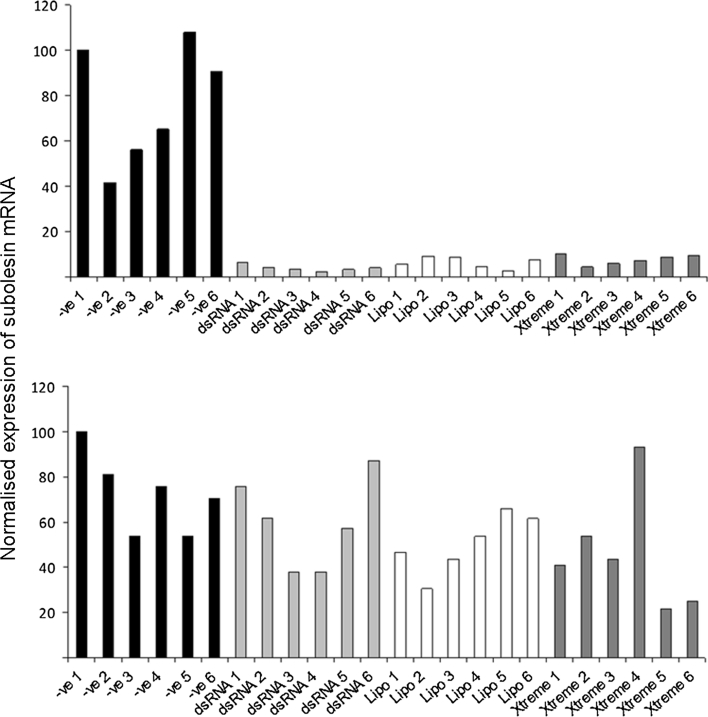
Fig. 4.
Efficiency of subolesin mRNA transcript knockdown in IDE8 and ISE6 cells. IDE8 (upper) and ISE6 (lower) cells were transfected (with or without a transfection reagent) with dsRNA targeting subolesin or negative control dsRNA targeting eGFP and incubated for 96 h. Data for all replicates are included to illustrate the level of variability encountered between individual cultures. Total RNA was extracted and equal amounts of RNA were used to make cDNA. Transcripts were measured by real-time PCR and Ct values for subolesin mRNA were then normalised against tick β-actin mRNA for each replicate. Subolesin mRNA expression levels (y-axis) are shown in arbitrary units. –ve dsRNA targeting eGFP, dsRNA only dsRNA targeting subolesin, lipo Lipofectamine 2000 + dsRNA targeting subolesin, x Xtreme + dsRNA targeting subolesin