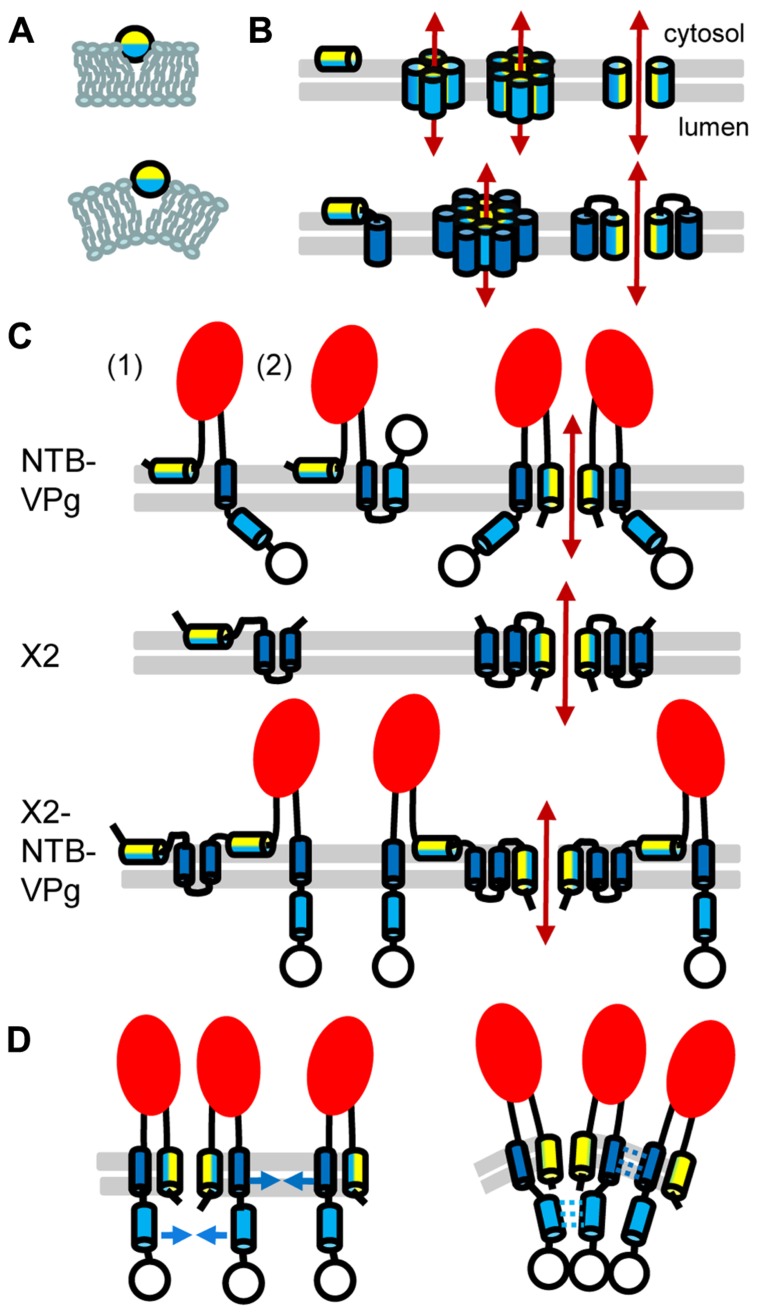
FIGURE 2.
Topology model for ToRSV membrane replication proteins. (A) Model for the parallel insertion of an amphipathic helix. The hydrophobic side of the helix (blue) inserts in one leaflet of the lipid bilayer while the polar/charged hydrophilic side of the helix (yellow) is exposed to the cytosolic face of the membrane. This insertion displaces the lipid headgroup, causing the acyl chain to reorient and inducing positive membrane curvature. (B) Model for the oligomerization of amphipathic helices and formation of an aqueous pore. In the top panel, an amphipathic helix is inserted parallel to the lipid bilayer (horizontal gray lines) of the membrane (left). Formation of an aqueous pore (double-ended red arrow) requires oligomerization of four or six amphipathic helices (middle). In the aqueous pore, the hydrophilic side of the helix (yellow) is exposed toward the pore, while its hydrophobic side (blue) is oriented toward the membrane lipid bilayer. A simplified representation of the pore shows only two molecules to better visualize each side of the amphipathic helix relative to the pore (right). In the bottom panel, a membrane protein consisting of an amphipathic helix and a hydrophobic helix (blue) is shown. After initial membrane insertion of the monomer with the amphipathic helix parallel to the membrane (left), an aqueous pore is formed by oligomerization of the amphipathic helix (middle). The hydrophobic helix of each molecule is located on the outside of the pore alongside the amphipathic helix (model shown for a hexamer). Hydrophobic interactions between the hydrophobic side of the amphipathic helix and the hydrophobic helix stabilize pore formation. A simplified representation of the pore shows only two molecules (right). (C) Predicted topologies for NTB–VPg, X2, and X2–NTB–VPg shown for monomers (left) or oligomers (right). Two possible topologies are shown for NTB–VPg monomers (1 and 2, see text). To simplify the figure, only two molecules are shown in the oligomer models. However, at least four molecules would be necessary to form an aqueous pore (as shown in B). The open circle represents the VPg domain and the red oval indicates the conserved NTB motif. (D) Model for the induction of positive membrane curvature by hydrophobic interactions of membrane proteins oligomers, shown for NTB–VPg. On the left, blue arrows represent possible hydrophobic interactions. These interactions (shown by broken blue lines on the right) would induce positive membrane curvature. Similar hydrophobic interactions are predicted to occur in X2 or X2–NTB–VPg oligomers (not shown).