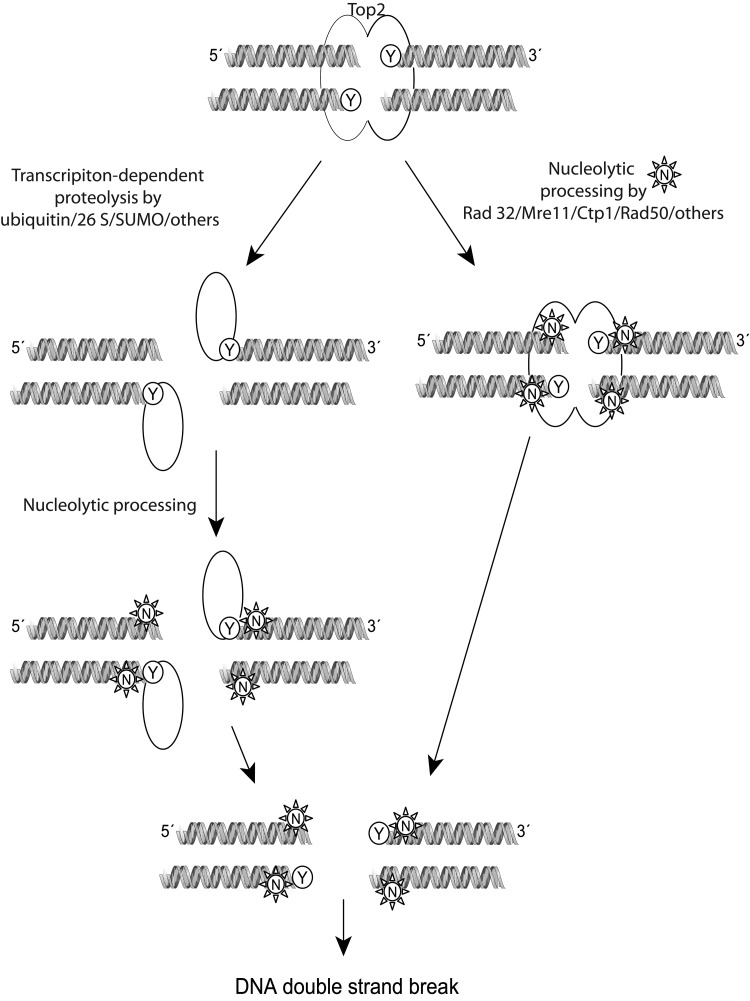
FIG. 5.
Mechanism of processing of the top2 cleavage complex with DNA by topoisomerase inhibitors. Either by encountering a replication fork or being sensed by RNA/DNA polymerases, the top2 covalent complexes are recognized and processed for repair by at least two pathways. Proteolytic processing of the top2 dimer could occur using the ubiquitin/26 S proteosome, or by SUMOylation. The enzyme is partially detached from the DNA but not completely removed. Other enzymes that are capable of cleaving remaining phosphotyrosyl bonds to DNA such as Tdp1/2 could assist in removal from DNA. Alternatively, nucleases such as members of the Rad family of nucleases can sever the link with DNA, generating a single- or double-stranded break. The single-stranded break could be converted to a double-stranded break during further processing or by encountering a replication fork. Homologous recombination or nonhomologous end joining can attempt to repair the double-strand breaks generated, but the exact mechanism is still being investigated. Modified with permission from John Nitiss (University of Illinois) (185). Tdp, tyrosyl DNA phosphodiesterase.