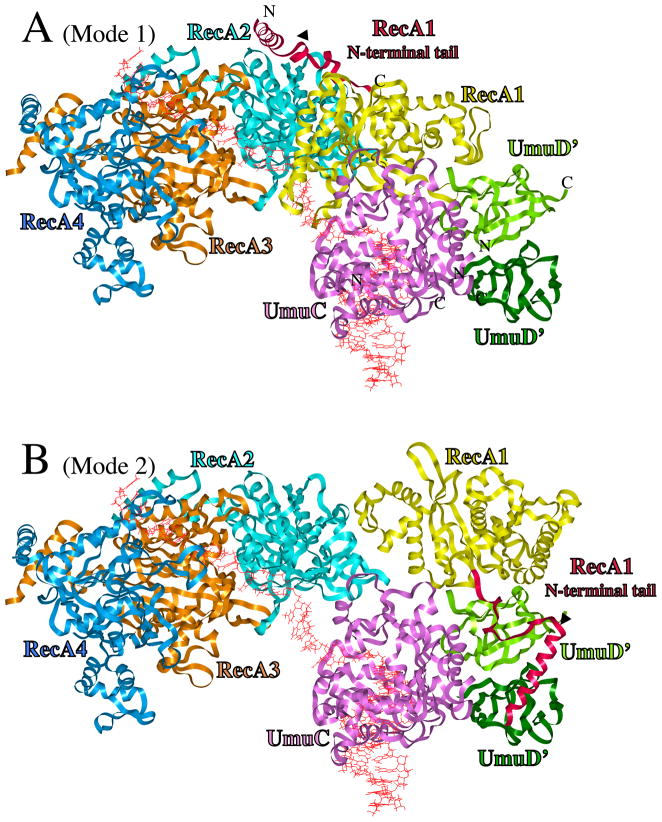
Figure 1.
UmuD′2C/RecA Models. (A) UmuD′2C is bound to the 3′-end of a RecA-filament, with RecA1 (yellow) primarily in contact with UmuC (Mode 1). From right-to-left, ribbons represent distal UmuD′ (dark green), proximal UmuD′ (green), UmuC (pink), RecA1 (yellow), RecA2 (turquoise), RecA3 (tan) and RecA4 (blue). The N-terminal 38 amino acids of RecA1 are highlighted in scarlet. DNA is red. (B) Structure for active UmuD′2C/RecA, in which RecA1 is rearranged to become primarily associated with UmuD′2 (Mode 2), thus freeing up three template nucleotides such that UmuC (pink) could initiate DNA synthesis. The N-terminal tail of UmuD′ (aa25-49), which is extended, is not shown. The C-terminus of UmuC (aa354-422), for which there is no model, is not present. The positioning of the N-terminus and C-terminus of RecA1, UmuC and both UmuD′s are indicated.