Figure 4.
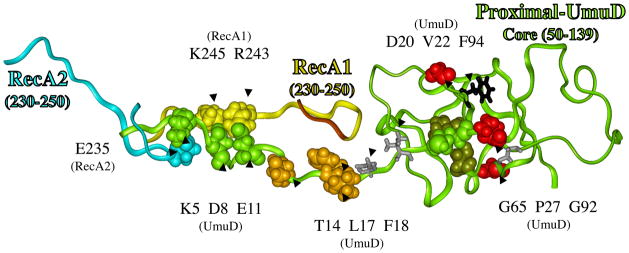
Structural model for how RecA might facilitate the cleavage of UmuD in its active site (red circle), where the catalytically important residue (S60/K97, dark green) cleave between amino acids C24 and G25 (green) to give UmuD′. A portion of RecA1 (aa230-250, yellow ribbon) and RecA2 (aa230-250, turquoise ribbon) are also shown. The N-terminal 49 amino acids of UmuD form a long tail, and evidence suggests it is in contact with core-UmuD (aa50-139) [84, 85]. Evidence also suggests that some portion of UmuD interacts with R243 in RecA to facilitate UmuD cleavage [86]. The N-terminal tail of UmuD is long enough and the spacing is reasonable to allow interactions involving: E11-UmuD with R243-RecA1; D8-UmuD with K245-RecA1; and K5-UmuD with E235-RecA2 E11. When these interactions are made, the cleavage site in UmuD (C24-G25, green) can sit in the proteolysis active site of UmuD (S60/K97, dark green), as indicated by the red oval. The figure also shows P27, G65 and G92 (red), which when mutated each give non-cleavable UmuD (see text). A UmuD triple mutant involving T14/L17/F18 (brown) is also non-cleavable (see text). The significance of D20 and V22 (gray sticks), along with F94 (black sticks) are discussed in the text.
