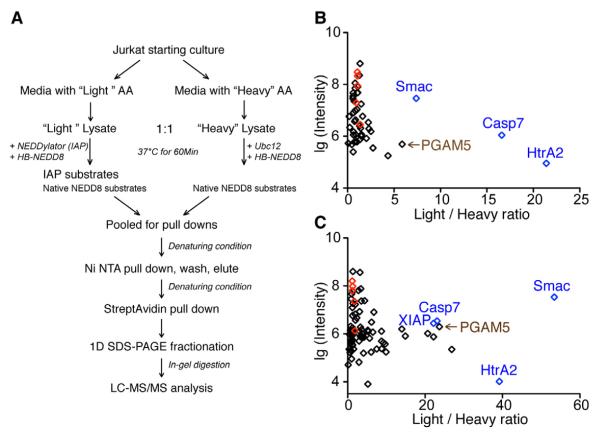
Figure 3. Identification of Potential XIAP and cIAP1 Substrates, Using Respective NEDDylators and SILAC-based Mass Spectrometry.
(A) Workflow for comparing proteins that are NEDDylated with (“Light”) or without (“Heavy”) the NEDDylator.
(B) Distribution of potential XIAP substrate protein Light/Heavy ratios obtained by the SILAC experiments. Each dot represents a protein, from which at least two unique peptides were identified. Light/Heavy ratios of each protein are plotted against the log scale of summed intensity. Known XIAP substrates caspase-7, HtrA2 and Smac are shown in blue; native NEDD8 substrate cullins, including Cul1, 2, 3, 4A, and 5 are shown in red.
(C) Distribution of potential cIAP1 substrate protein Light/Heavy ratios obtained by a similar SILAC experiment. Labels are the same as in (B).