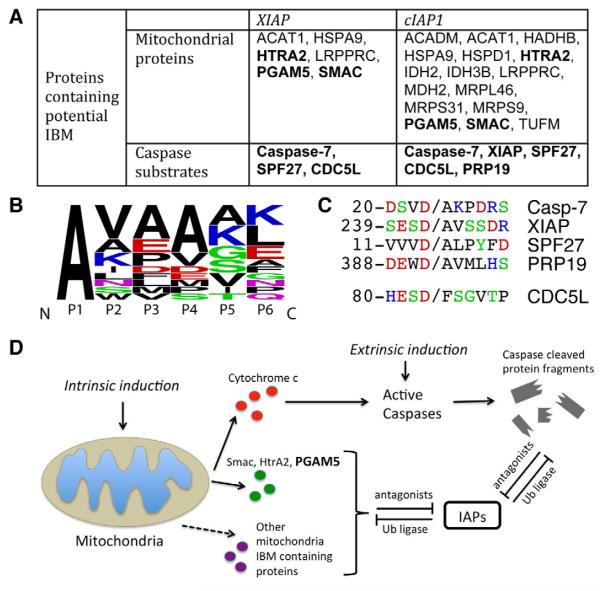
Figure 6. Proteolytic Cleavage Generates a N-terminal IAP Binding Motif (IBM) that is a Common Feature of IAP Substrates.
(A) Summary table of potential XIAP and cIAP1 substrates containing IBM-like sequences from combined data sets. For simplicity, gene names are used to represent each protein. The full protein names are listed in Table S4.
(B) Sequence logo representation of the frequency of amino acids (Crooks et al., 2004) in the IBM-like cleavage sites identified in 16 mitochondrial proteins listed in table (A). Amino acids are colored according to their chemical properties.
(C) Sequence alignment of amino acid residues surrounding the caspase cleavage sites. Caspases cleave at Asp and generate IBM-like sequences. (D) A model for IAP-IBM interactions.
See Table S4