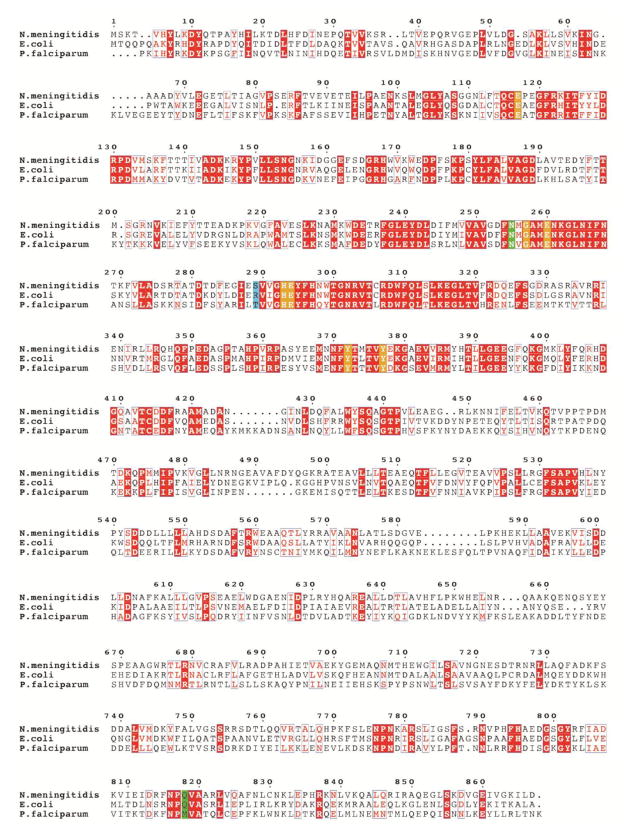
Fig. 6.
Multiple sequence alignment of three alanine aminopeptidases from bacteria (E. coli, N. meningitidis) and protozoan (P. falciparum, truncated to residues 195–1085, lacking the N-terminal extension, which contains 3 asparagine-rich regions and a putative transmembrane domain). Sequence identities are highlighted in red and similarities are shown as red letters. The key active site residues involved in the coordination of zinc atoms and in the catalytic mechanism are highlighted in orange. The residues, which are located in the S1 pocket and can control its specificity, are shown in green. The residues equivalent to Ser289 of Nm (involved in binding of the Tyr P1′ residue) are highlighted in blue (see the text for further discussion).