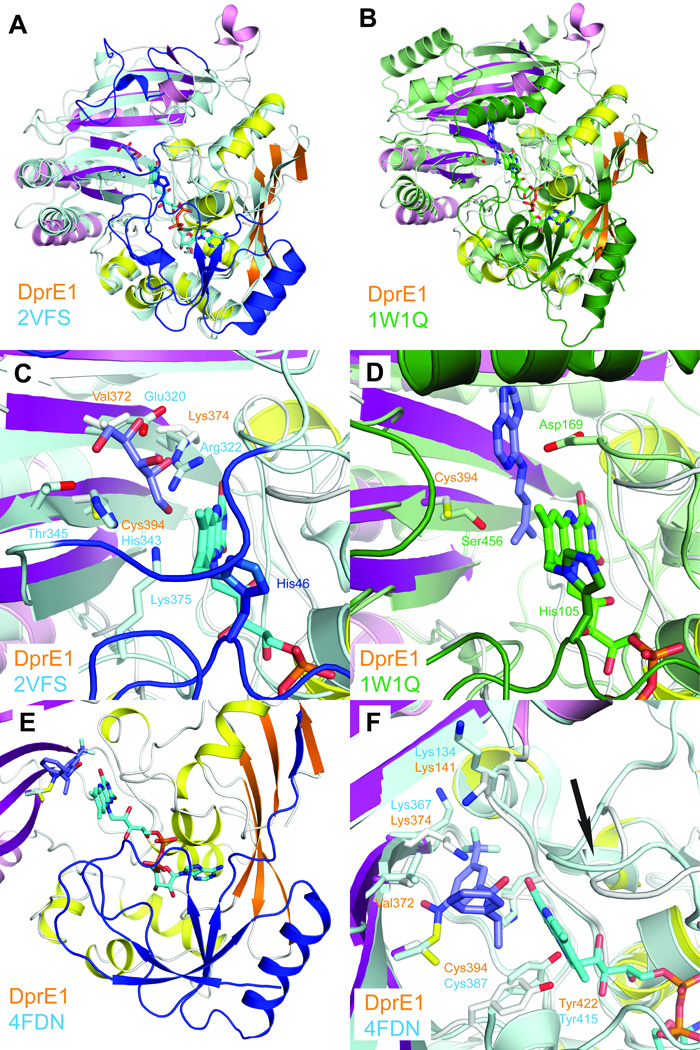
Figure 2. Comparison of DprE1 with structurally related enzymes.
a. Least-squares superposition of DprE1 with alditol oxidase (colored in cyan and blue, PDB entry 2VFS). N-terminal residues absent in the DprE1 structure and residues approximately equivalent to the flexible loop regions are colored in blue.
b. Least-squares superposition of DprE1 with cytokinin dehydrogenase (colored in green, PDB entry 1W1Q). N-terminal residues absent in the DprE1 structure and residues approximately equivalent to the flexible loop regions are colored in dark green.
c. Close-up view comparing the active site region of DprE1 with alditol oxidase. FAD is shown in cyan sticks bound to His46. The substrate xylitol is shown in purple sticks and xylitol-coordinating residues are shown in cyan sticks. Residues approximately equivalent to the flexible loop regions are colored in blue.
d. Close-up view comparing the active site region of DprE1 with cytokinin dehydrogenase. FAD is shown in green sticks bound to His105. The cytokinin dehydrogenase product N6-isopentenyladenine is shown in purple sticks. Residues approximately equivalent to the flexible loop regions are colored in dark green.
e. N-terminal region absent in the DprE1 structure as observed in the DprE1 structure from M. tuberculosis (colored in blue, PDB code 4FDN). FAD and the DprE1 inhibitor CT325 in DprE1_tb are shown in cyan and blue sticks, respectively. The structure of DprE1_sm is colored as before.
f. Comparison of residues in the DprE1 active site (shown in white sticks) with DprE1 from M. tuberculosis (cyan sticks). The conformational change of the active site loop between residues 121 and 128 is indicated with an arrow.