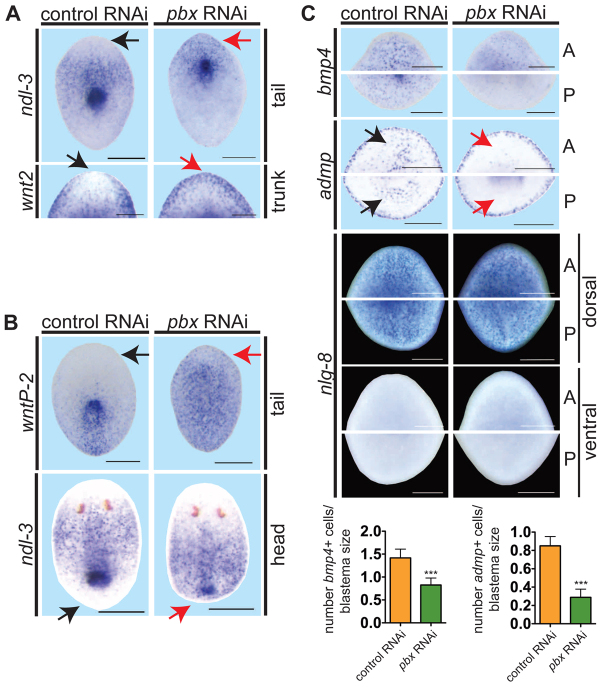
Fig. 3.
pbx(RNAi) animals display abnormal pole gene expression and fail to re-scale expression gradients during regeneration. (A-C) Whole-mount ISH of regenerating pieces of planarians at day 6. Anterior is to the top. Black arrows, normal gene expression location (control); red arrows, aberrant expression in pbx(RNAi) animals. (A) pbx(RNAi) animals exhibited aberrant anterior blastema gene expression: ndl-3 (control, n=15/15; pbx RNAi, n=1/16 display restriction from the head tip of tail fragments) and wnt2 (control, n=10/11; pbx RNAi, n=0/13 display restriction from the head tip; head blastema of trunk fragment shown). (B) pbx(RNAi) animals failed to re-scale the wntP-2 expression gradient (control, n=17/18; pbx RNAi, n=4/20 tail fragments show re-scaling) and ndl-3 expression (control, n=22/25; pbx RNAi, n=1/22 head fragments show re-scaling). (C) Dorsal bmp4 expression (control, n=18; pbx RNAi, n=20) and ventral admp expression (control, n=13; pbx RNAi, n=10) were reduced in pbx(RNAi) animals, but dorsal expression of nlg-8 was normal in both anterior (A) and posterior (P) blastemas (control, n=13/13; pbx RNAi, n=12/12). ‘Head’, ‘trunk’ and ‘tail’ refer to a head fragment regenerating a tail, a fragment with head and tail amputated transversely, and a tail fragment regenerating a new head after transverse amputation, respectively. Graphs show quantification of bmp4+ and admp+ cells normalized to blastema size (mean±s.e.m.; ***P=0.0,001, t-test). Scale bars: 200 μm.