Table 1.
Structures of fluorescent adenosine receptor (AR) ligands. In this table, the conjugates are structurally divided into three moieties to fulfill pharmacological, covalent linking, and fluorescent functions (see Figure 1 for structures of fluorophores and Table 2 for references).
| No. | Compound Namea | Pharmacophore | Linker | Fluorophore |
|---|---|---|---|---|
| 1 | FITC-ADAC | 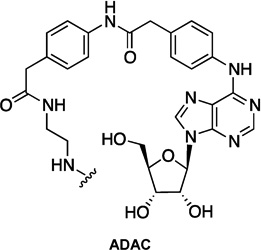 |
- | FITC |
| 2 | NBD-ADAC | NBD | ||
| 3 | FITC-APEC | 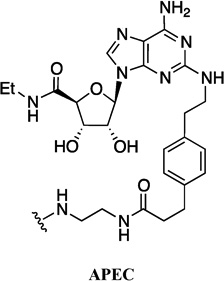 |
- | FITC |
| 4 | Alexa488- APEC (salt form) |
Alexa Fluor 488 5/6 mixed isomers |
||
| 5 | MRS5424 | Alexa Fluor 532 | ||
| 6 | Dansyl-NECA (n=3) |
 |
(CH2)3NH | Dansyl |
| 7 | Dansyl-NECA (n=4)b |
(CH2)4NH | ||
| 8 | Dansyl-NECA (n=6) |
(CH2)6NH | ||
| 9 | Dansyl-NECA (n=8) |
(CH2)8NH | ||
| 10 | Dansyl-NECA (n=10) |
(CH2)10NH | ||
| 11 | Dansyl-NECA (n=12) |
(CH2)12NH | ||
| 12 | ABEA-AO-dansyl (8) |
(CH2)4NHCO(CH2)7NH | ||
| 13 | AUEA-dansyl (10) | (CH2)11NH | ||
| 14 | NBD-NECA (n=2) | (CH2)2NH | NBD | |
| 15 | NBD-NECA (n=4) | (CH2)4NH | ||
| 16 | NBD-NECA (n=6) | (CH2)6NH | ||
| 17 | NBD-NECA (n=8) | (CH2)8NH | ||
| 18 | NBD-NECA (n=10) | (CH2)10NH | ||
| 19 | APrEA-X-BY630 | (CH2)3NH | Bodipy 630/650-X | |
| 20 | ABEA-X-BY630 | (CH2)4NH | ||
| 21 | APEA-X-BY630 | (CH2)5NH | ||
| 22 | AOEA-X-BY630 | (CH2)8NH | ||
| 23 | ADOEA-X-BY630 | ((CH2)2O)2(CH2)2NH | ||
| 24 | NECA-Bodipy- 630/650 (n=2) |
(CH2)2NH | Bodipy-630/650 | |
| 25 | NECA-Bodipy- 630/650 (n=3) |
(CH2)3NH | ||
| 26 | NECA-Bodipy- 630/650 (n=4) |
(CH2)4NH | ||
| 27 | NECA-Bodipy- 630/650 (n=8) |
(CH2)8NH | ||
| 28 | NECA-Bodipy- 630/650 (n=11) |
(CH2)11NH | ||
| 29 | NECA-Bodipy- 630/650 (PEG n=2) |
CH2(CH2OCH2)2CH2NH | ||
| 30 | NECA-Bodipy- 630/650 (PEG n=3) |
CH2(CH2OCH2)3CH2NH | ||
| 31 | ABEA-BYLF | (CH2)4NH | Bodipy FL | |
| 32 | ABEA-X-BYLF | (CH2)4NHCO(CH2)5NH | ||
| 33 | ABEA-X-Texas Red |
(CH2)4NH | Texas Red | |
| 34 | ABEA-Cy5 | Cy5 | ||
| 35 | ABEA-EVOBlue30 | EVOBlue 30 | ||
| 36 | ABA-X-BY630 |  |
(CH2)4NH | Bodipy 630/650-X |
| 37 | FITC-XAC |  |
- | FITC |
| 38 | FITC-Gly3-XAC | CONHCH2 | ||
| 39 | XAC-X-BY630 | - | Bodipy 630/650-X | |
| 40 | XAC-X-Texas Red | - | Texas Red | |
| 41 | XAC-Cy5 | Cy5 | ||
| 42 | XAC-AEAO-BYFL | CO(CH2)6CONH(CH2)2NH | Bodipy FL | |
| 43 | XAC-EVOBlue30 | - | EVOBlue 30 | |
| 44 | XAC-Dansyl | Dansyl | ||
| 45 | XAC-AO-Dansyl | CO(CH2)7NH | ||
| 46 | XAC-AHH-Dansyl | CO(CH2)5NHCO(CH2)5NH | ||
| 47 | TQO-C8-X- BODIPY630 |
 |
CO(CH2)2CONH(CH2)8NH | Bodipy 630/650-X |
| 48 | TQO-PEG-X- BODIPY630 |
CO(CH2)2CONH((CH2)2O)2(CH2)2NH | ||
| 49 | TQO-PEG-X- BODIPY-TR |
Bodipy-TR | ||
| 50 | TQO-PEG-Cy5 | Cy5 | ||
| 51 | TQO-PEG-TAMRA | Tamra 5/6-X mixed isomers |
||
| 52 | TQO-extPEG- TAMRA |
5-Tamra-PEG | ||
| 53 | MRS5346 |  |
CH2CONH(CH2)2NH | Alexa Fluor 488 5-isomer |
| 54 | MRS5418 | CH2CONH((CH2)2NH)2 | Bodipy650/665-X | |
| 55 | MRS5347 | CH2CONH(CH2)2 | Tamra | |
| 56 | MRS5449 |  |
CO(CH2)3 | Alexa Fluor 488 azide 5-isomer |
| 57 | PTP-FITC (1) |  |
NH | FITC |
| 58 | PTP-FITC (2) | (CH2)3NH | ||
| 59 | PTP-FITC (3) | (CH2)4NH | ||
| 60 | PTP-FITC (4) | (CH2)5NH | ||
| 61 | PTP-FITC (5) | ((CH2)2O)2(CH2)2NH | ||
| 62 | PTP-FITC (6) | (CH2)3O((CH2)2O)2(CH2)3NH | ||
| 63 | MRS5218 | 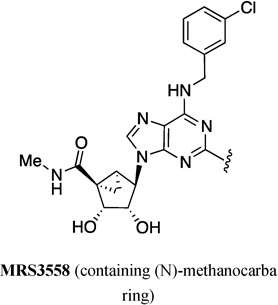 |
C≡C(CH2)2CONH(CH2)2NH | Cy5 |
| 64 | MRS5243 | C≡C(CH2)4 | Squaraine- Rotaxane |
|
| 65 | MRS5238 | C≡C(CH2)4 | Alexa Fluor 488 azide |
|
| 66 | MRS5704 | C≡C | 4-Pyrene | |
| 67 | MRS5783 | C≡C | 1-Pyrene | |
| 68 | MRS5421 |  |
||
| 69 | MRS5422 | |||
| 70 | MRS5397 | 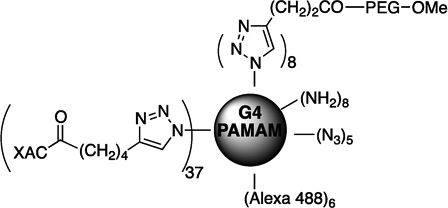 |
||
| 71 | MRS5399 | 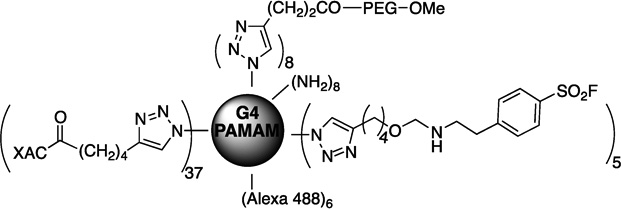 |
||
| 72 | MRS5303 |  |
||
abbreviations: ABA: N6-(aminobutyl)adenosine; ABEA: N6-(4-aminobutyl)-5′-ethylamino-5′-oxo-5′-deoxyadenosine; ADAC: adenosine amine congener; ADOEA: N6-(8-amino-3,6-dioxaoctyl)-5′-ethylamino-5′-oxo-5′-deoxyadenosine; AEAO: 8-(2-aminoethylamino)-8-oxooctanoyl; AHH: 6-(6-aminohexanamido)hexanoyl; AO: 8-aminooctanoyl; AOEA: N6-(8-aminooctyl)-5′-ethylamino-5′-oxo-5′-deoxyadenosine; APEA: N6-(5-aminopentyl)-5′-ethylamino-5′-oxo-5′-deoxyadenosine; APrEA: N6-(3-aminopropyl)-5′-ethylamino-5′-oxo-5′-deoxyadenosine; APEC: 2-[(2-aminoethylamino)carbonylethyl-phenylethylamino]-5′-ethylcarboxamidoadenosine; AUEA: N6-(11-aminoundecyl)-5′-ethylamino-5′-oxo-5′-deoxyadenosine; CGS15943: 9-chloro-2-(2-furanyl)-[1,2,4]triazolo[1,5-c]quinazolin-5-amine; (N)-methanocarba: [3.1.0]bicyclohexane; NECA: 5'-N-ethylcarboxamidoadenosine; PAMAM: polyamidoamine; PTP: pyrazolo[4,3-e][1,2,4]triazolo[1,5-c]pyrimidin-5-amine; QD: quantum dot; SCH442416: 2-(2-furanyl)-7-[3-(4-methoxyphenyl)propyl]-7H-pyrazolo[4,3-e][1,2,4]triazolo[1,5-c]pyrimidin-5-amine; TQO: [1,2,4]-triazolo[4,3-a]quinoxalin-1-one; TQZ: [1,2,4]-triazolo[1,5-c]quinazolin-5-amine; XAC: xanthine amine congener.
structure is same as ABEA-dansyl.36
