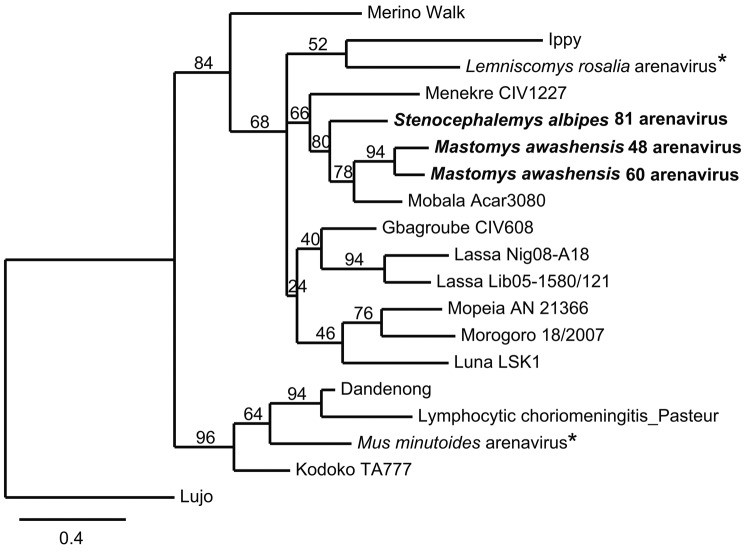
Figure 1.
Maximum-likelihood tree of Old World arenaviruses showing the position of 3 arenaviruses (boldface; GenBank accession nos. JQ956481–JQ956483) found in kidney samples of Awash multimammate mice (Mastomys awashensis) and Ethiopian white-footed mice (Stenocephalemys albipes). The tree was constructed on the basis of analysis of partial sequences of the RNA polymerase gene; phylogeny was estimated by using the maximum-likelihood method with the GTR + I + Γ (4 rate categories) substitution model to account for rate heterogeneity across sites as implemented in the PhyML program (8). Lujo arenavirus was used as an outgroup. Numbers represent percentage bootstrap support (1,000 replicates). *Arenaviruses from Tanzania that have not yet been named (9). Scale bar indicates nucleotide substitutions per site. GenBank accession numbers of the virus strains: EU136039, GU830849, AY363902, EF179864, GU979511, GU481071, DQ868486, GU182412, FJ952385, AB586645, GU830863, GU078661, DQ328876, AY363904, EU914110, GU182413.