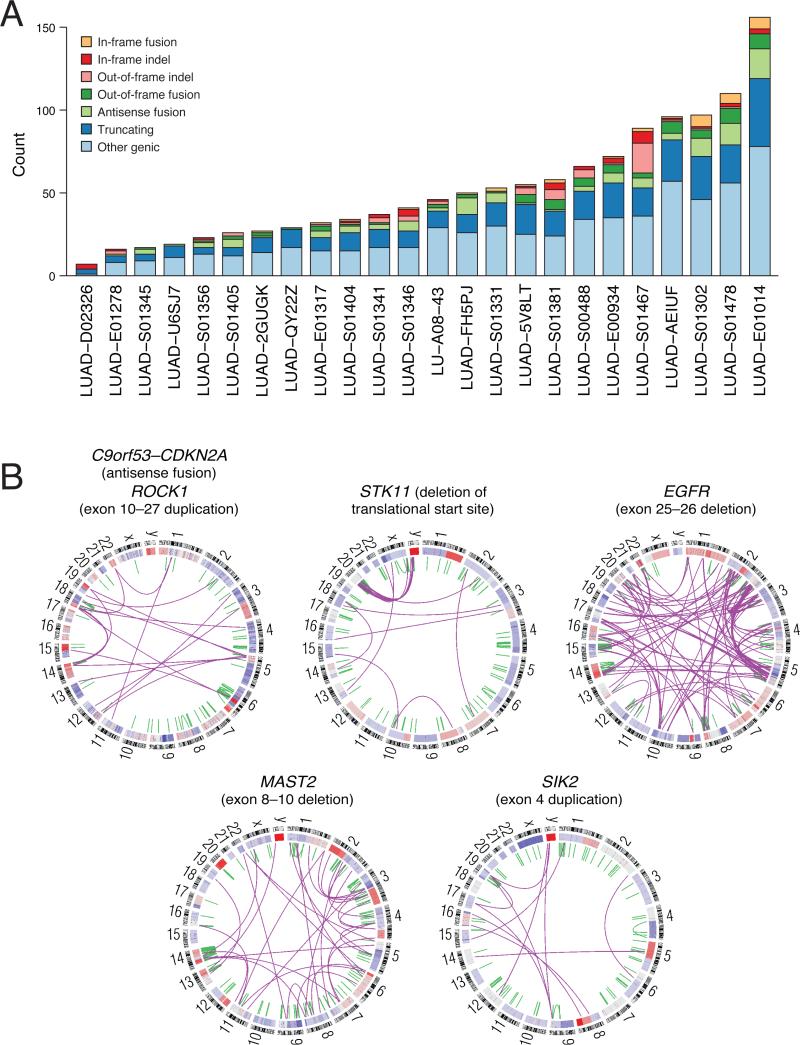
Figure 4. Whole genome sequencing of lung adenocarcinoma.
(A) Summary of genic rearrangement types across 25 lung adenocarcinoma whole genomes. Stacked-bar plot depicting the types of somatic rearrangement found in annotated genes by analysis of whole genome sequence data from 25 tumor/normal pairs. The “Other Genic” category refers to rearrangements linking an intergenic region to the 3’ portion of a genic footprint. (B) Representative Circos (Krzywinski et al., 2009) plots of whole genome sequence data with rearrangements targeting known lung adenocarcinoma genes CDKN2A, STK11 and EGFR and novel genes MAST2, SIK2, and ROCK1. Chromosomes are arranged circularly end-to-end with each chromosome's cytobands marked in the outer ring. The inner ring displays copy number data inferred from whole genome sequencing with intrachromosomal events in green and interchromosomal translocations in purple.