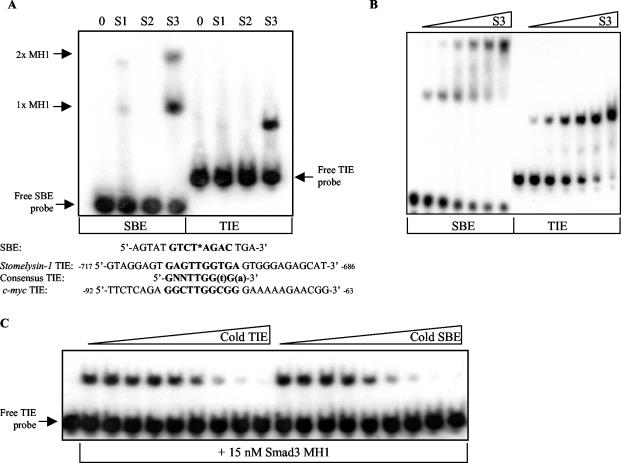
FIG. 4.
The MH1 domain of Smad3 specifically and directly binds to the c-myc TIE with a relative affinity similar to that of the SBE. (A) A 15 nM concentration of recombinant Smad1 (S1), Smad2 (S2), or Smad3 (S3) MH1 domain was individually incubated with radiolabeled SBE and c-myc TIE-containing probes, and an EMSA was performed as described in Materials and Methods. Two Smad3 MH1 complexes shifted the 16-bp SBE probe due to the incorporated palindromic double site, as depicted in bold. One molecule of the MH1 domain bound to one molecule of SBE is represented by 1x MH1, whereas two molecules of the MH1 domain bound to one molecule of DNA are represented by 2x MH1. The employed 30-bp c-myc TIE probe, equivalent to nucleotides of the c-myc promoter −92 to −63 relative to the P2 transcriptional start site, is depicted below. Sequence encompassing the first identified TIE within the rat stromelysin promoter as well as the composition of the subsequently proposed “consensus” TIE are listed below. Nucleotides in bold represent invariance, whereas lowercase letters signify preferred nucleotides. (B) The relative affinities of the Smad3 MH1-SBE and Smad3-TIE interactions were determined by EMSA, in which increasing concentrations of Smad3 MH1 recombinant protein (S3) were incubated with a fixed amount of radiolabeled SBE or c-myc TIE probe. Threefold serial dilutions of protein, from 556 to 2.29 nM, were used as indicated. The free or unbound probes are the lowest bands depicted within their respective lanes. (C) The similarity of the relative affinities of Smad3 MH1 for SBE and c-myc TIE probes was confirmed by a DNA cold competitor EMSA as described in Materials and Methods. Fifteen nanomolar Smad3 MH1 domain recombinant protein was incubated with radiolabeled c-myc TIE and increasing amounts of cold competitor DNA. The EMSA samples incubated with unlabeled c-myc TIE oligonucleotide are shown on the left, whereas those with unlabeled SBE are on the right. Threefold dilutions of DNA, from 700 to 0.320 nM, were used as indicated. All the data depicted are representative of at least three independent experiments.