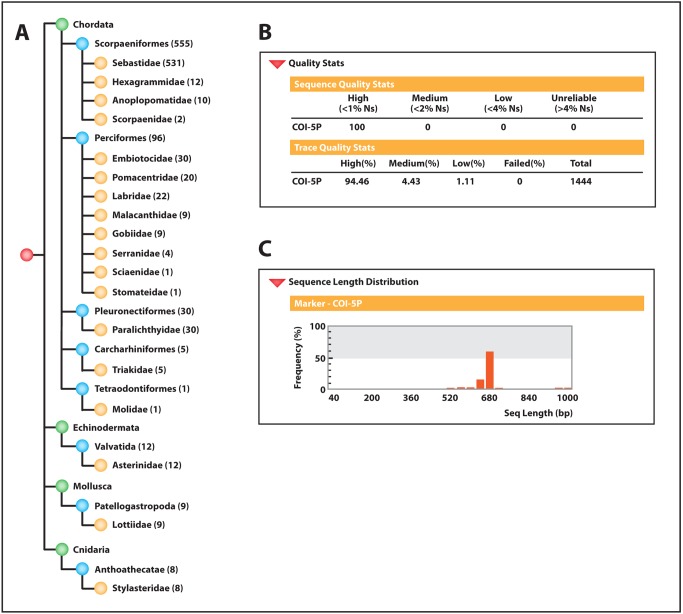
Figure 5. Reference DNA barcode records generated by project participants.
(A) As of October 2012, students and teachers have generated and submitted complete reference DNA barcode records from 716 unique individuals representing four animal phyla, eight orders, 18 families, 26 genera (not shown), and 53 species (not shown). 716 records are currently published in GenBank (see Text S2 for the accession numbers corresponding to each published record). GenBank accession numbers, specimen and collection data, nucleotide sequences, trace files, and primer details are also available within the Barcoding Life's Matrix project folder, which is accessible through the BOLD Public Data Portal (http://www.boldsystems.org/index.php/Public_BINSearch?searchtype=records; use search term “BLM”). (B) Quality statistics for edited and unedited CO1 nucleotide sequence data. CO1 sequences edited by project participants from raw trace files contain no ambiguous base calls (Ns), stop codons, contaminating sequences, or insertions or deletions. Each amplicon was sequenced bidirectionally to yield at least one forward and one reverse trace file for each barcode record. Of the 1,444 trace files generated, 94.46% are categorized as high quality (mean Phred quality score >40 [28]), 4.43% are categorized as medium quality (mean Phred quality score = 30–40), and 1.11% are categorized as low quality (mean Phred quality score <30). (C) Nucleotide length distribution of CO1 sequences generated for each specimen. All 716 sequences generated by project participants exceeded the minimum barcode length of 500 nucleotides, with a minimum sequence length of 502 nucleotides, a maximum sequence length of 1,152 nucleotides, and a mean sequence length of 720 nucleotides.