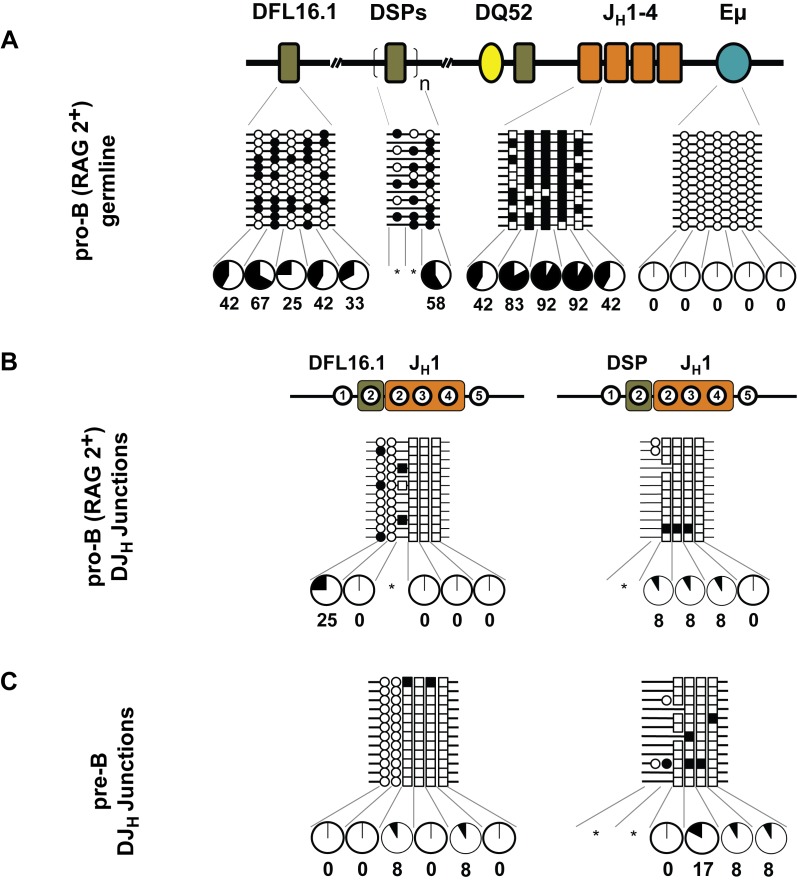
Figure 4. DNA methylation status after the first step of IgH locus recombination.
Pro-B cells were purified from the bone marrow of wild-type C57BL/6 mice. Data shown are derived from two independent preparations of pro- and pre-B cells obtained from six to eight mice in each experiment. This cell population contains a mix of germline and partially rearranged IgH alleles. After bisulfite modification, the genomic DNA was used to amplify unrearranged (A) and DJH rearranged junctions containing DFL16.1 and DSP gene segments (B). Cytosines derived from DH and JH gene segments are marked as circles and squares, respectively. Filled and open circles, or squares, indicate methylated and unmethylated cytosines, respectively. Numbers within regions marked as DFL16.1, DSP and JH1 in (B) denote CpG dinucleotides corresponding to the configuration of these residues at the respective unrearranged gene segments. For example, of the five CpGs at unrearranged DFL16.1 only the first two are retained in DFL16.1/JH1 junctions. The total numbers of CpG dinucleotides are reduced in junctional sequences because some residues are lost during VDJ recombination as described in the text. Additional heterogeneity is due to the imprecise nature of recombination. Pie charts summarize the percentage of methylated cytosines at each position, except where the number of alleles falls below 12 (indicated by asterisks). (C) Methylation state of recombined DJH alleles in purified pre-B cells. This population contains a mix of VDJH recombined and DJH recombined IgH alleles (see Figure 5A). Note that the number of circles and squares representing DH- or JH-associated CpGs differed in the DJH junctions compared to the corresponding unrearranged regions due to junctional variability.