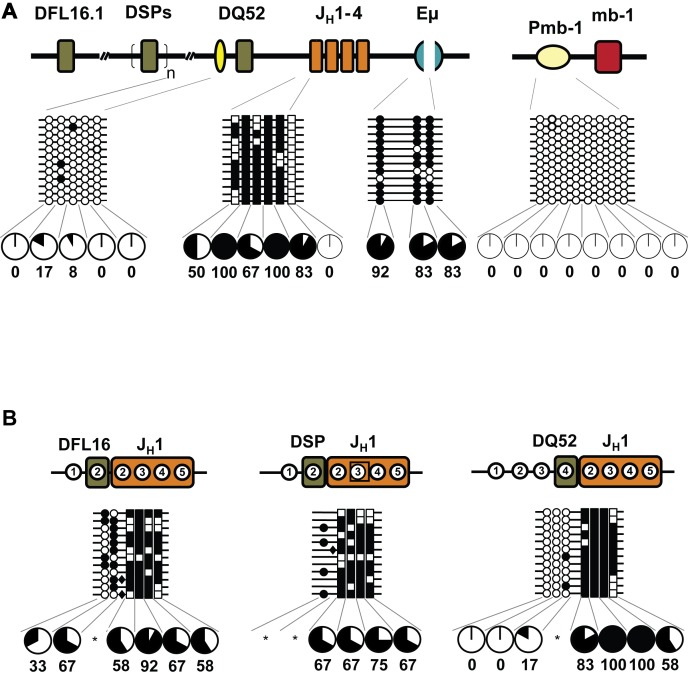
Figure 6. DNA methylation state of IgH alleles in the absence of the intronic enhancer Eμ.
Pro-B cells were purified from the bone marrow of Eμ-deficient mice [40]. Genomic DNA was subjected to bisulfite modification followed by PCR amplification of germline gene segments (A) and DJH junctions (B). The 220-bp deletion of Eμ is represented as partial blue arcs separated by a gap. Filled and open circles, or squares, indicate methylated and unmethylated cytosines, respectively. Numbers within regions marked as DFL16.1, DSP, and JH1 denote CpG dinucleotides corresponding to the configuration at the respective unrearranged gene segments. For example, of the five CpGs at unrearranged DFL16.1 only the first two are retained in DFL16.1/JH1 junctions. Three out of five cytosines used to analyze wild-type alleles remain in this deletion, and their methylation status is shown immediately below the disrupted enhancer. Sequences that substitute for the enhancer also contain CpGs whose methylation status is shown in Figure S5. (B) Recombined DFL16.1, DSP and DQ52 to JH1 junctions were amplified, cloned, and sequenced. Untemplated CpGs incorporated during VDJ recombination were found to be methylated (filled diamonds). Data shown are derived from two independent preparations of pro-B cells from Eμ-deleted mice, with four to six mice in each experiment.