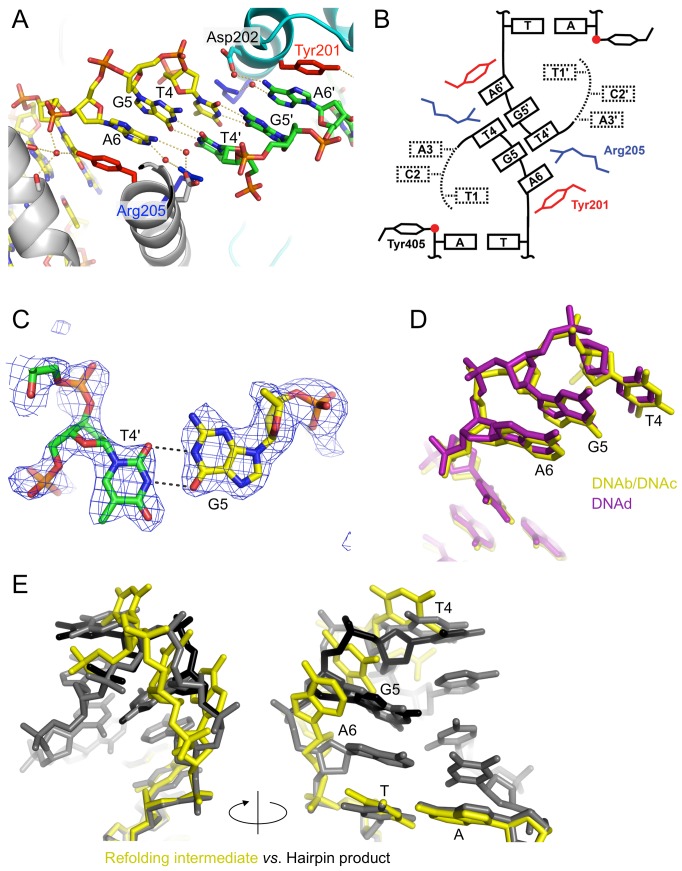
Figure 7. The strand-refolding intermediate.
(A) The open DNA conformation observed in the phosphotyrosine complexes stabilized by stacking of the flipped-out bases, G-T wobble base-pairs, and water-mediated hydrogen bonds. The protein and DNA molecules are colored as in Figure 4A. (B) Schematic diagram of the strand-refolding intermediate DNA conformation. The 5′ bases Thy1, Cyt2, and Ade3 are flexible. Thy1 in our crystal structures is either missing or replaced by cytosine to block ligation and trap the phosphotyrosine bond. (C) Simulated annealing omit Fo-Fc electron density contoured at 3.0 σ for the G-T wobble pair. (D) 5′-overhang conformation observed in the phosphotyrosine complexes (the refolding intermediate) trapped using two different types of suicide DNA substrates. The structure obtained with the nicked suicide substrate (DNAb/DNAc) is shown in yellow, while that obtained with a mismatch-based suicide substrate (DNAd) is shown in purple. The tri-nucleotide stretch Thy4, Gua5, and Ade6 is superimposable with an r.m.s.d. of 0.78 Å. The deviation comes mostly from the phosphate backbone atoms. (E) Comparison of the DNA structures between the refolding intermediate (yellow) and the hairpin telomere product (alternate conformations shown in black and grey). The superposition is based on the double-stranded stem region of the DNAs.