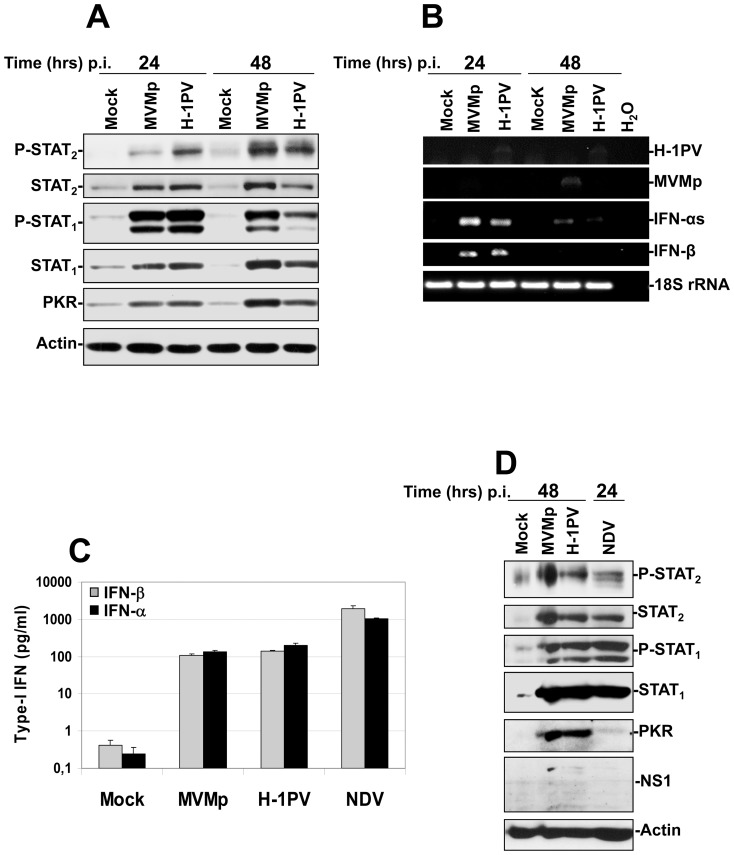
Figure 6. Activation of both IFN-producing and IFN-signaling pathways in hPBMCs upon parvovirus infection and comparison of their intensity to that triggered by NDV.
(A, B and D) hPBMCs collected from the blood of healthy donors were distributed into 6-well plates at 1×107 cells/5 ml culture medium/well. They were then mock-treated or infected with MVMp or H-1PV at 20 PFUs/cell or with NDV (6 HU/106 cells). Cultures were harvested in PBS and centrifuged at the time p.i. indicated in each figure. (A, D) One half of each pellet was resuspended in Ripa buffer supplemented with phosphatase and protease inhibitors in order to perform Western blot experiments whereas (B) total RNAs were extracted from the rest of each cell pellet using the RNeasy kit. (A, D) Total proteins were extracted from each sample as described in Materials and Methods. Seventy µg total proteins per sample were then subjected to 10% SDS-PAGE, transferred onto membranes, and probed with antibodies specific for total and phosphorylated STAT2 and STAT1 polypeptides as well as for PKR. Actin was used as an internal loading control. Each presented blot is representative of 4 additional which gave similar results. (B). One µg of isolated total RNA was then reverse transcribed into cDNA and 10% of this product was subjected to PCR reactions using sets of primers specific to each indicated mRNA. PCR product of 18S ribosomal RNA was used as housekeeping gene to normalize loading. No signal was detected in the samples when omitting the reverse transcriptase. Presented data are representative of 4 experiments which all gave similar results. (C) hPBMCs collected from the blood of healthy donors were distributed into 24-well plates at 1×106 cells/500 µl culture medium/well. They were then mock-treated or infected with MVMp, H-1PV (20 PFUs/cell) or NDV (6 HU/106 cells). After a period of incubation of 24 hrs media were collected, centrifuged in order to discard cellular debris and analyzed by Enzyme-linked Immuno-Sorbent Assay (ELISA) for their content in IFN-α and IFN-β. Results are expressed as means+standard deviations of seven independent experiments.